CASMI: And the Winner is . . .
Abstract
:1. Introduction
1.1. Background of the Inaugural CASMI
- 20/05/2012: Public release of www.casmi-contest.org
- 20/07/2012: Public release of challenge data
- 31/01/2013: Deadline for submissions (extended to 05/02/2013)
- 06/02/2013: Public release of solutions
- 22/02/2013: Public release of automatic evaluation (07/03/2013 for resubmissions)
2. Methods: Evaluation and Ranking of Participants
2.1. Absolute Ranking
2.2. Relative Ranking

- −
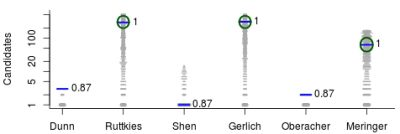
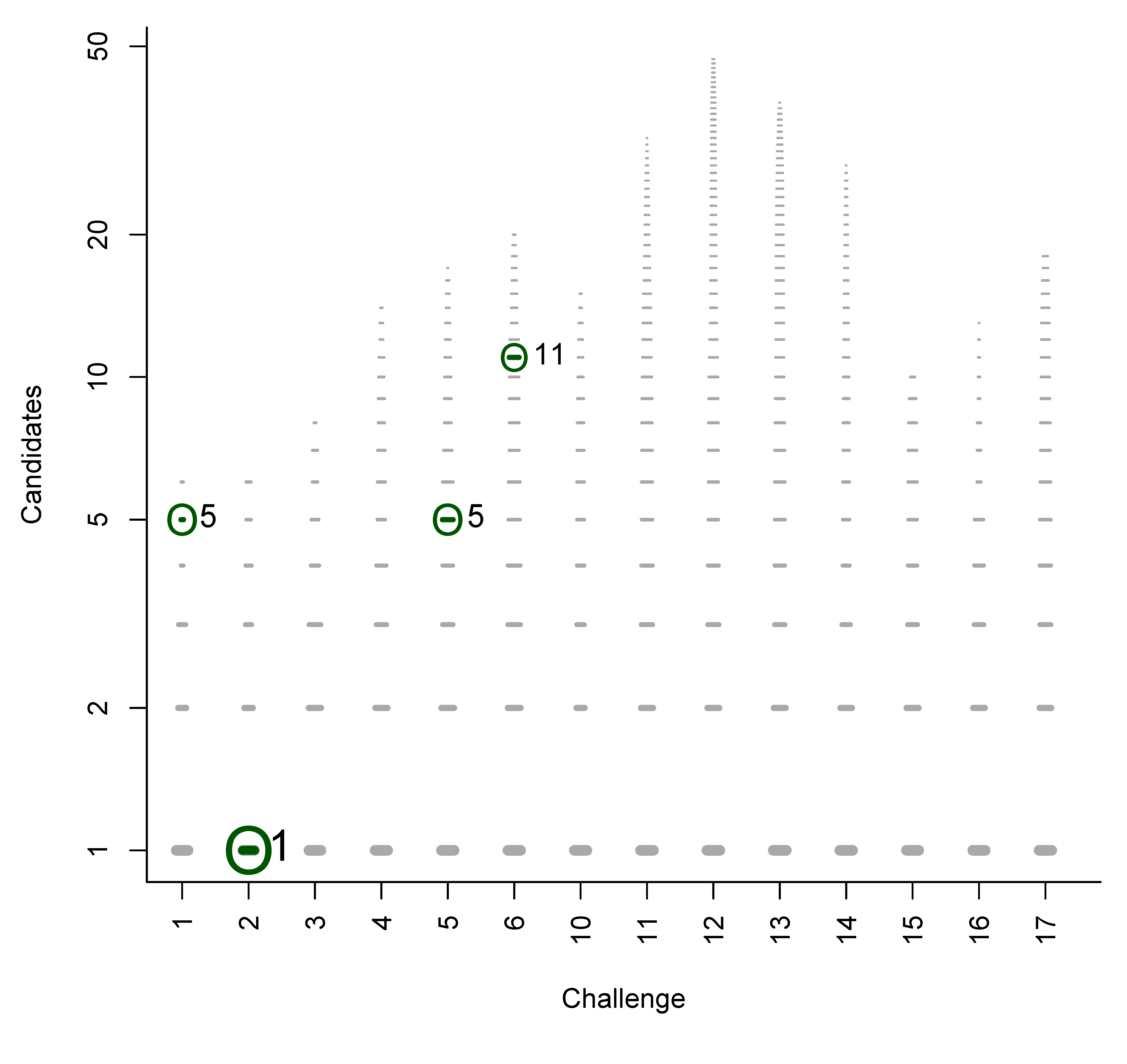
- For Challenge 1, Category 2, Shen et al. have TC = 6, BC = 4, WC = 1 and a RRP of 0.20; while Ruttkies et al. have TC = 1423, BC = 21, EC = 24, WC = 1378 and a much higher RRP = 0.98, although the absolute rank of 45 is worse than Shen et al.’s 5;
- −
- For Challenge 1, Category 2, Oberacher and Dunn et al. have TC = 1 and the RRP is undefined (both were correct), while Gerlich et al. have TC = 1356, BC = 0, WC = 1355, with RRP = 1.00; here Oberacher, Dunn et al. and Gerlich et al. share the honours, which is not visible from the RRP.
- −
- For Challenge 12, Category 2, all participants missed the correct answer; the RRP is undefined.
2.3. Normalised Scores and Weighted RRP
 in a submission such that ∑is̃i = 1 and calculated a weighted RRP using the score s̃j of the correct solution to obtain the sum of all better-scoring candidates
in a submission such that ∑is̃i = 1 and calculated a weighted RRP using the score s̃j of the correct solution to obtain the sum of all better-scoring candidates  and equivalently
and equivalently  to give:
to give:
- −
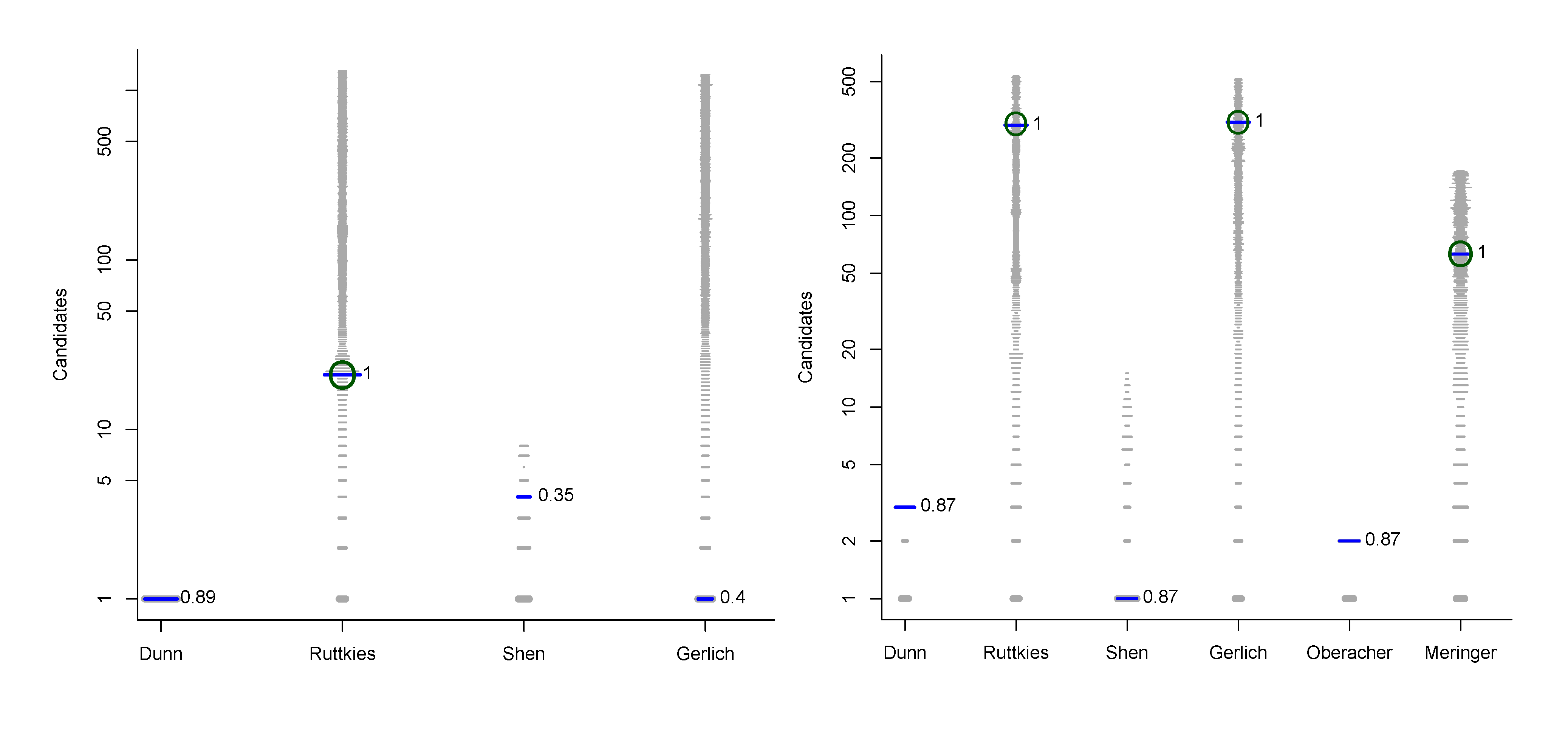
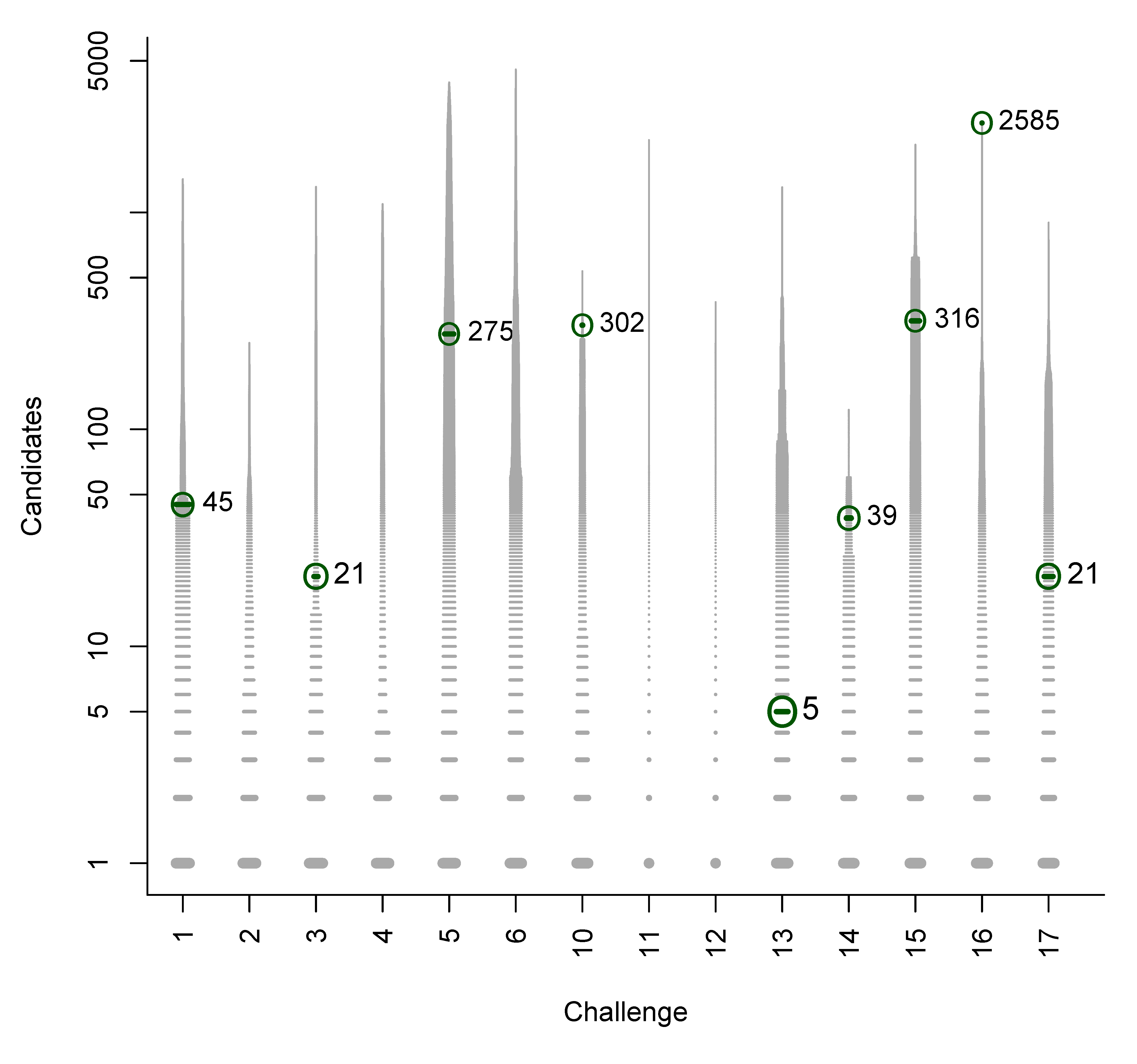
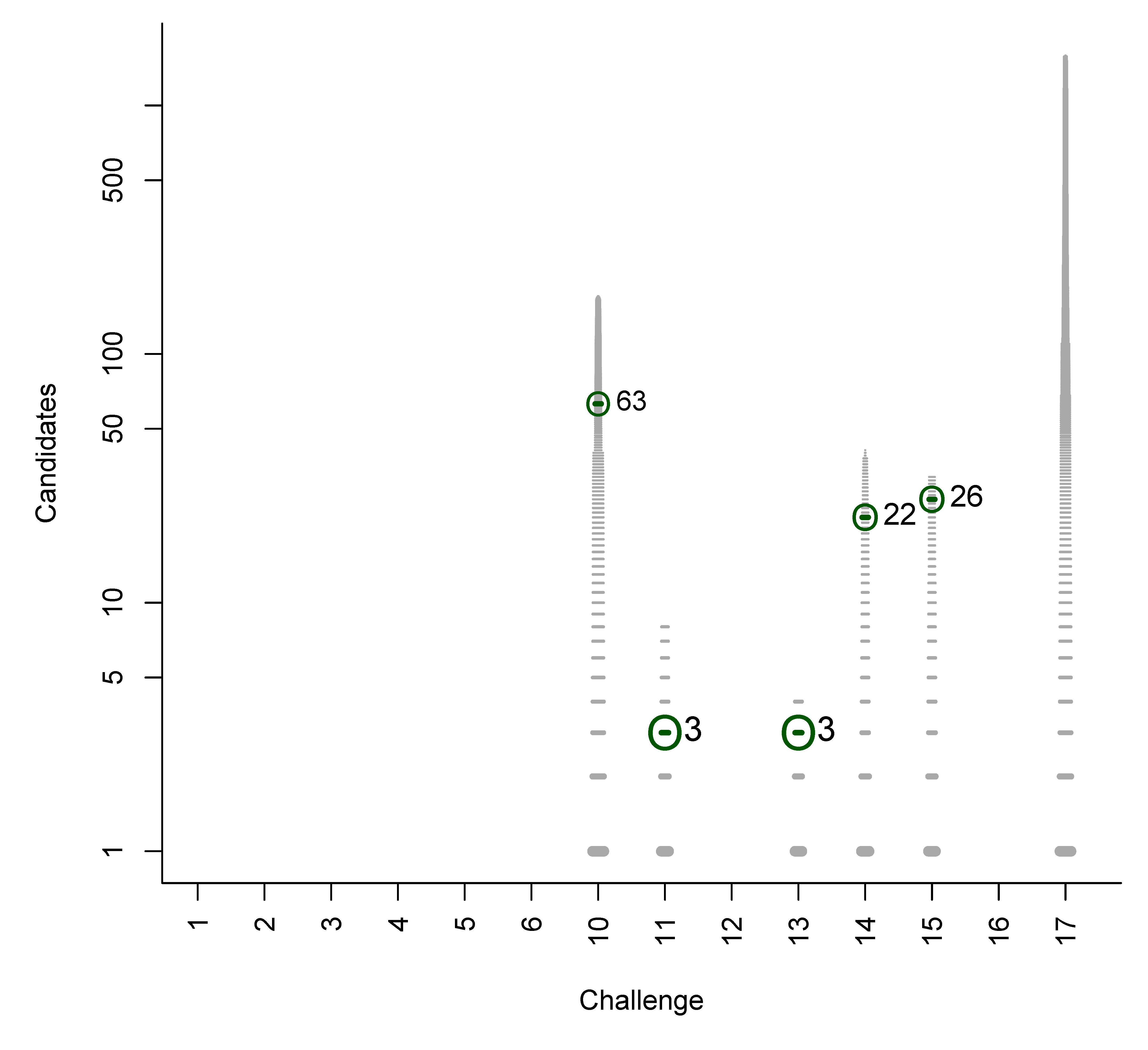
- For Challenge 10, Category 2, Ruttkies et al., Gerlich et al. and Meringer et al. had the correct solution with absolute ranks 302, 307 and 63, respectively. The wRRPs of 0.01, 0.35 and 0.49 show that the scoring used by the latter two were more useful than the first in selecting the correct candidate, however given normalised scores (s̃) of ≤ 0.01 for all three, few users would have considered these candidates;
- −
- For Challenge 2, Category 2, Shen et al. and Gerlich et al. have wRRPs of 1.00 and 0.91 respectively.
- −
- For Challenge 13, Category 1, Dührkop et al., Neumann et al. and Meringer et al. all have rank = 1, RRP = 1.00 and wRRP = 1.00, with TC = 20, 141 and 10, respectively. The normalised scores of 1.00, 1.00 and 0.14, respectively, indicate that the scoring system of Dührkop et al. and Neumann et al. weight the top candidate heavily—which is advantageouswhen the top candidate is correct, but can offer a false sense of security when interpreting the results.
2.4. Similarity Between Submissions and the Correct Solution (Category 2)

- −
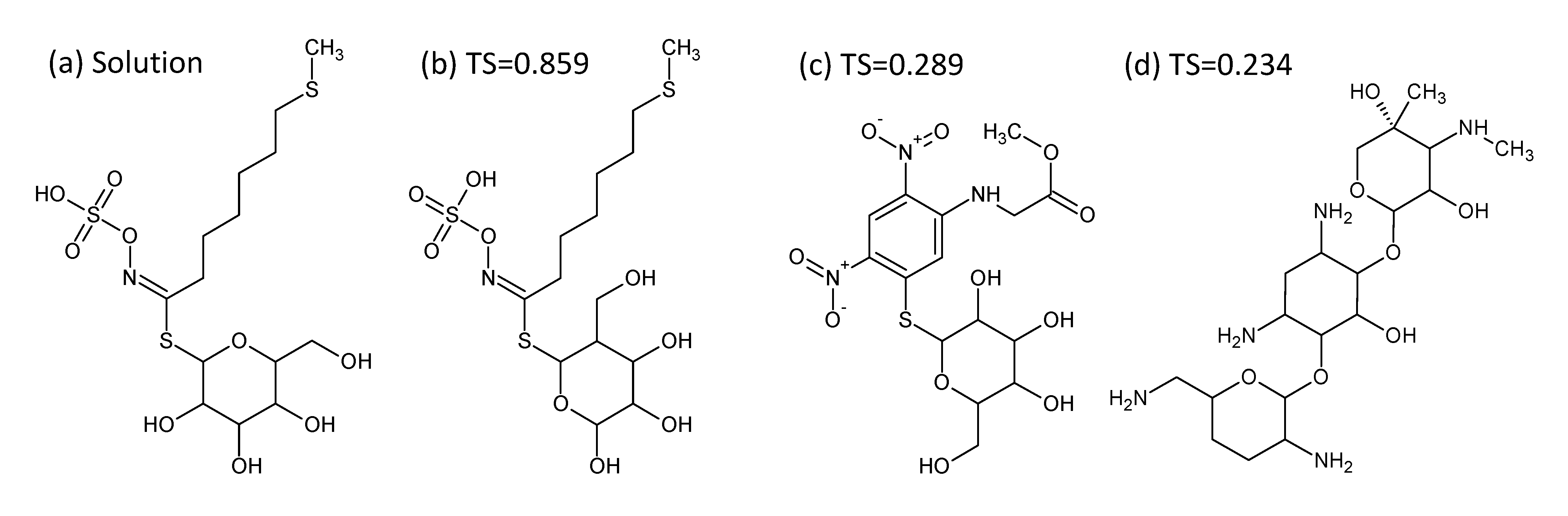
- In Challenge 3, Category 2, the entry from Ruttkies et al. contained the correct compound, while the other three contestants in this category missed it. The correct answer and TS with the most similar entries from the other participants are shown in Figure 2. This figure and the similarity scores show that Dunn et al. were the closest of the three contestants that missed.
- −
- In Challenge 10, Category 2, Dunn et al., Shen et al. and Oberacher all missed the correct solution, 1-aminoanthraquinone. However, all entries contained the positional isomer 2-aminoanthraquinone, in third, first and second place (by score), respectively. The Tanimoto similarity between the two positional isomers is 0.842.
2.5. Evaluation Framework
3. Results by Participant
3.1. Participation Rate
3.2. Summary Results by Participant
| Participant | Subm. | Cor(win) | Avg. Rank | Avg. TC | Avg. BC | Avg. RRP | Avg. wRRP | Avg. s̃ |
|---|---|---|---|---|---|---|---|---|
| Dunn et al. | 11 | 9(9) | 1.11 | 1.4 | 0.1 | 0.500 | 0.926 | 0.889 |
| Shen et al. | 14 | 8(3) | 2.88 | 11.1 | 1.9 | 0.719 | 0.670 | 0.168 |
| Dührkop et al. | 14 | 8(5) | 2.25 | 128.8 | 1.3 | 0.992 | 0.625 | 0.602 |
| Dührkop et al. r | 14 | 12 | 5.75 | 134.7 | 4.7 | 0.883 | 0.423 | 0.406 |
| Neumann et al. | 13 | 9(5) | 4.33 | 639.1 | 3.3 | 0.991 | 0.565 | 0.561 |
| Neumann et al. r | 14 | 12 | 4.83 | 1915.0 | 3.8 | 0.997 | 0.508 | 0.434 |
| Meringer et al. | 14 | 11(9) | 4.45 | 34.4 | 2.7 | 0.847 | 0.759 | 0.275 |
| Meringer et al. r | 14 | 14 | 3.29 | 36.6 | 1.7 | 0.941 | 0.885 | 0.226 |
| Participant | Subm. | Cor(win) | Avg. Rank | Avg. TC | Avg. BC | Avg. RRP | Avg. wRRP | Avg. s̃ |
|---|---|---|---|---|---|---|---|---|
| Dunn et al. | 11 | 3(2) | 5.7 | 4.7 | 3.3 | 0.556 | 0.606 | 0.4294 |
| Ruttkies et al. | 14 | 9(2) | 401.0 | 1618.9 | 138.4 | 0.813 | 0.547 | 0.0041 |
| Ruttkies et al. r | 14 | 14 | 319.7 | 1226.3 | 188.1 | 0.838 | 0.616 | 0.0069 |
| Shen et al. | 14 | 4(2) | 5.5 | 19.4 | 4.3 | 0.614 | 0.520 | 0.1226 |
| Gerlich et al. | 14 | 11(5) | 237.5 | 1631.2 | 236.5 | 0.882 | 0.864 | 0.0020 |
| Gerlich et al. r | 14 | 14 | 305.4 | 2878.1 | 304.3 | 0.873 | 0.862 | 0.0010 |
| Oberacher | 5 | 3(3) | 1.0 | 1.2 | 0.0 | − | 1.000 | 1.0000 |
| Meringer et al. | 6 | 5(2) | 23.4 | 307.7 | 22.4 | 0.470 | 0.457 | 0.0887 |
| Meringer et al. r | 6 | 6 | 29.2 | 258.5 | 28.2 | 0.551 | 0.535 | 0.0741 |
3.3. External Participants
3.4. Internal Participants
4. Results by Challenge
4.1. Statistics for Category 1
| Chal. | Subm. | Correct | Avg. Rank | Avg. TC | Avg. BC | Avg. RRP | Avg. wRRP | Avg. s̃ |
|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 7.4 | 906.0 | 6.4 | 0.761 | 0.388 | 0.221 |
| 2 | 5 | 4 | 1.3 | 2931.2 | 0.3 | 1.000 | 0.764 | 0.228 |
| 3 | 5 | 4 | 10.5 | 802.0 | 9.5 | 0.944 | 0.455 | 0.253 |
| 4 | 5 | 5 | 1.8 | 143.6 | 0.8 | 0.958 | 0.546 | 0.175 |
| 5 | 5 | 5 | 7.6 | 142.8 | 6.4 | 0.679 | 0.505 | 0.250 |
| 6 | 5 | 5 | 2.0 | 319.0 | 1.0 | 0.755 | 0.605 | 0.236 |
| 10 | 5 | 5 | 1.0 | 29.4 | 0.0 | 1.000 | 1.000 | 0.718 |
| 11 | 4 | 1 | 4.0 | 53.3 | 0.0 | 0.700 | 0.400 | 0.200 |
| 12 | 4 | 3 | 10.0 | 229.5 | 8.0 | 0.850 | 0.213 | 0.042 |
| 13 | 5 | 3 | 1.0 | 89.8 | 0.0 | 1.000 | 1.000 | 0.714 |
| 14 | 5 | 5 | 1.0 | 10.8 | 0.0 | 1.000 | 1.000 | 0.801 |
| 15 | 5 | 5 | 2.0 | 18.0 | 1.0 | 0.797 | 0.675 | 0.540 |
| 16 | 4 | 1 | 4.0 | 294.8 | 1.0 | 0.895 | 0.749 | 0.082 |
| 17 | 5 | 4 | 1.0 | 21.4 | 0.0 | 1.000 | 1.000 | 0.857 |
4.2. Statistics for Category 2
| Chal | Subm. | Correct | Avg. Rank | Avg. TC | Avg. BC | Avg. RRP | Avg. wRRP | Avg. s̃ |
|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 2.4 | 646.2 | 1.4 | 0.732 | 0.818 | 0.4166 |
| 2 | 4 | 3 | 2.7 | 221.5 | 1.7 | 0.996 | 0.975 | 0.1012 |
| 3 | 4 | 2 | 11.5 | 1012.0 | 10.5 | 0.994 | 0.931 | 0.0033 |
| 4 | 4 | 2 | 264.0 | 1617.3 | 263.0 | 0.890 | 0.741 | 0.0006 |
| 5 | 4 | 4 | 668.3 | 1798.0 | 664.3 | 0.630 | 0.472 | 0.0842 |
| 6 | 4 | 3 | 105.7 | 2014.5 | 104.3 | 0.773 | 0.641 | 0.0190 |
| 10 | 6 | 3 | 434.7 | 287.2 | 427.3 | 0.391 | 0.190 | 0.0022 |
| 11 | 4 | 3 | 65.3 | 487.3 | 56.7 | 0.858 | 0.816 | 0.0458 |
| 12 | 3 | 2 | 86.0 | 1783.3 | 83.5 | 0.971 | 0.815 | 0.0045 |
| 13 | 6 | 4 | 2.5 | 736.5 | 0.8 | 0.777 | 0.865 | 0.3149 |
| 14 | 6 | 4 | 16.0 | 121.7 | 13.5 | 0.723 | 0.565 | 0.0227 |
| 15 | 6 | 4 | 50.3 | 877.2 | 48.5 | 0.700 | 0.712 | 0.2579 |
| 16 | 3 | 2 | 1649.5 | 2129.3 | 766.0 | 0.574 | 0.340 | 0.0001 |
| 17 | 5 | 3 | 53.7 | 727.4 | 52.7 | 0.960 | 0.911 | 0.0035 |
5. Discussion
6. Conclusions and Perspectives
Conflict of Interest
Acknowledgements
Appendix
A. Summary of CASMI 2012 Challenges
| Challenge | Trivial Name | Formula |
|---|---|---|
| 1 | Kanamycin A | C18H36N4O11 |
| 2 | 1,2-Bis-O-sinapoyl-beta-d-glucoside | C28H32O14 |
| 3 | Glucolesquerellin | C14H27NO9S3 |
| 4 | Escholtzine | C19H17NO4 |
| 5 | Reticuline | C19H23NO4 |
| 6 | Rheadine | C21H21NO6 |
| 10 | 1-Aminoanthraquinone | C14H9NO2 |
| 11 | 1-Pyrenemethanol | C17H12O |
| 12 | alpha-(o-Nitro-p-tolylazo)acetoacetanilide | C17H16N4O4 |
| 13 | Benzyldiphenylphosphine oxide | C19H17OP |
| 14 | 1H-Benz[g]indole | C12H9N |
| 15 | 1-Isopropyl-5-methyl-1H-indole-2,3-dione | C12H13NO2 |
| 16 | 1-[(4-Methoxyphenyl)amino]-1-oxo-2-propanyl-6-oxo-1-propyl-1,6-dihydro-3-pyridazinecarboxylate | C18H21N3O5 |
| 17 | Nitrin | C13H13N3 |
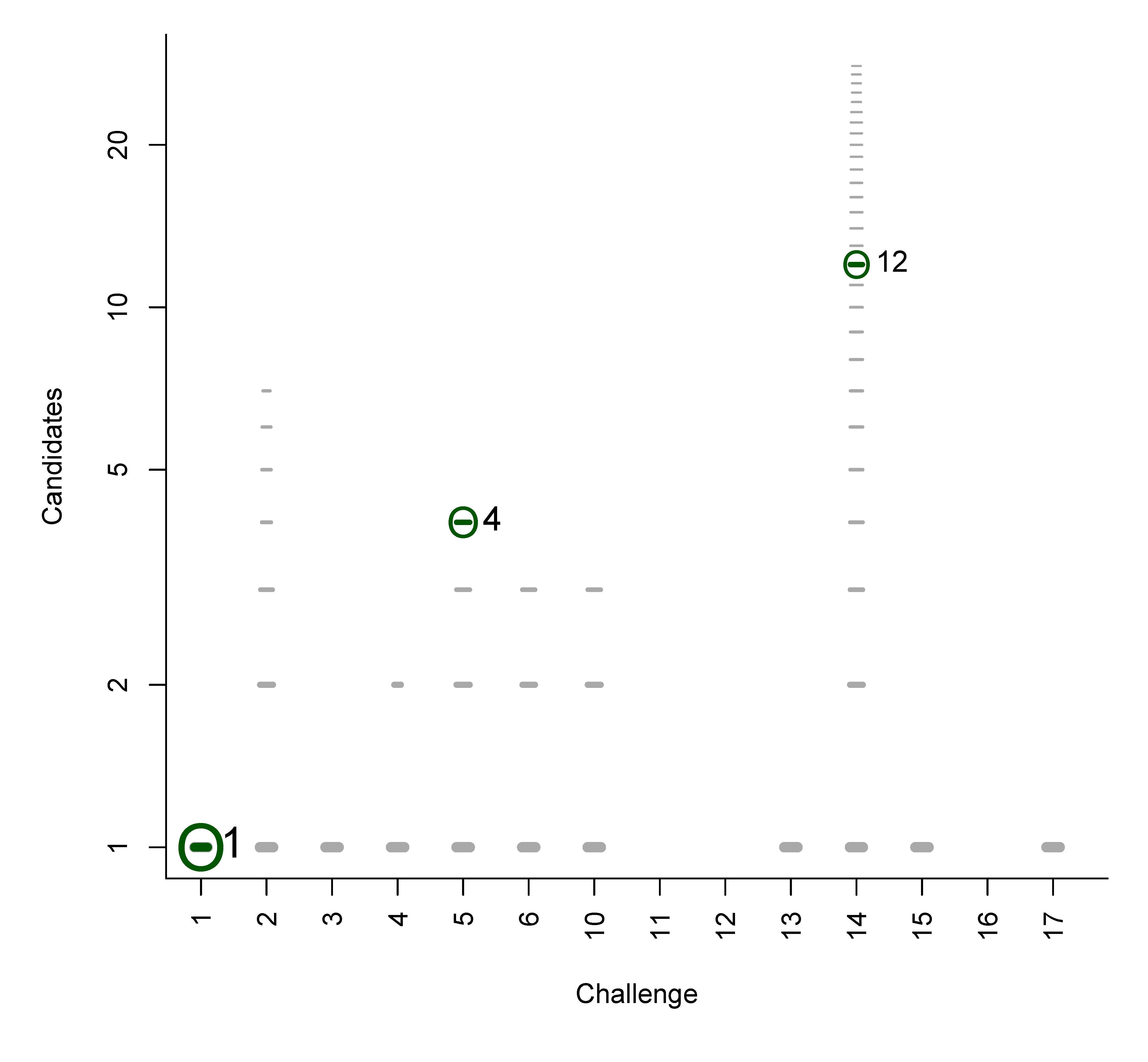
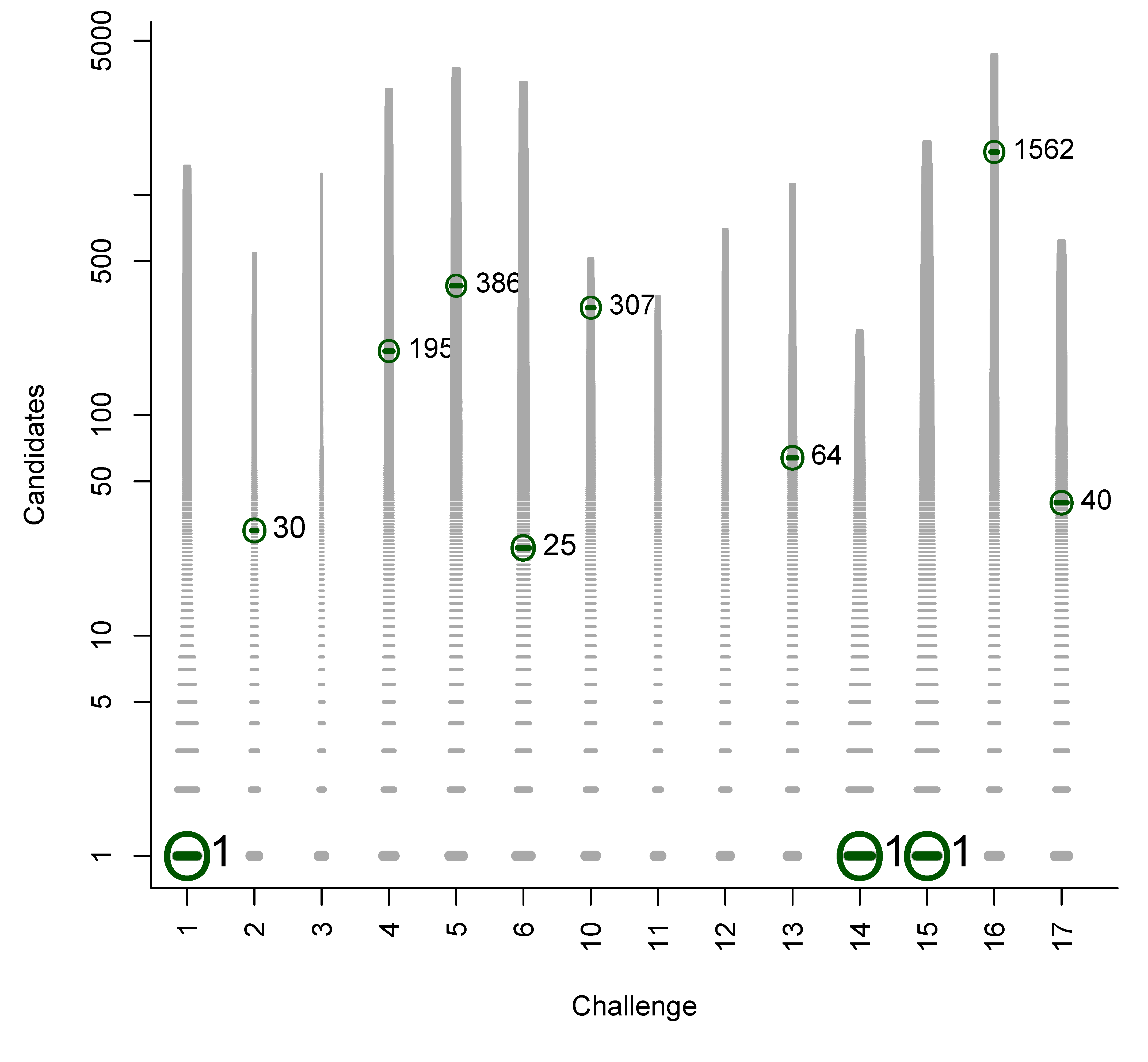
B. Participant Submissions by Score
References
- PSPC. Protein Structure Prediction Center. Available online: http://predictioncenter.org/ (accessed on 11 March 2013).
- Moult, J.; Fidelis, K.; Kryshtafovych, A.; Tramontano, A. Critical assessment of methods of protein structure prediction (CASP) Round IX. Proteins Struc. Funct. Bioinforma. 2011, 79, 1–5. [Google Scholar]
- Schymanski, E.L.; Neumann, S. Critical assessment of small molecule identification contest. 2012. Available online: http://casmi-contest.org/ (accessed on 20 May 2013).
- Schymanski, E.L.; Neumann, S. CASMI: Challenges and solutions. Metabolites 2013. sumitted. [Google Scholar]
- Schymanski, E.L.; Neumann, S. Critical Assessment of small molecule identification contest news archive. 2012. Available online: http://casmi-contest.org/archive.shtml/ (accessed on 20 May 2013).
- Schymanski, E.L.; Neumann, S. Critical assessment of small molecule identification: A new contest series. 2012. Available online: http://www.metabonews.ca/Oct2012/MetaboNews_-Oct2012.htm\#spotlight/ (accessed on 11 March 2013).
- Pervukhin, A.; Neumann, S. Rdisop: Decomposition of Isotopic Patterns. 2013. Available online: http://www.bioconductor.org/packages/devel/bioc/html/Rdisop.html/ (accessed on 12 March 2013).
- Böcker, S.; Letzel, M.; Lipták, Zs.; Pervukhin, A. Decomposing Metabolomic Isotope Patterns. In Proceedings of Workshop on Algorithms in Bioinformatics (WABI 2006), Zurich, Switzerland, 11–13 September 2006; Volume 4175, pp. 12–23.
- Heller, S.R.; McNaught, A.D. The IUPAC International Chemical Identifier (InChI). Chemistry International 2009, 31, 1, ISSN 0193-6484. [Google Scholar]
- IUPAC. The IUPAC International Chemical Identifier, International Union of Pure and Applied Chemistry. 2012. Available online: http://www.iupac.org/inchi/ (accessed on 11 March 2013).
- Daylight. SMILES-A Simplified Chemical Language. Daylight Chemical Information Systems Inc., 2012. Available online: http://www.daylight.com/dayhtml/doc/theory/theory.smiles.html (accessed on 11 March 2013).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [Google Scholar] [CrossRef]
- Pletnev, I.; Erin, A.; McNaught, A.; Blinov, K.; Tchekhovskoi, D.; Heller, S. InChIKey collision resistance: An experimental testing. J. Cheminf. 2012, 4, 39. [Google Scholar] [CrossRef]
- Gerlich, M.; Neumann, S. MetFusion: Integration of compound identification strategies. J. Mass Spectrom. 2013, 48, 291–298. [Google Scholar] [CrossRef]
- Steinbeck, C.; Han, Y.; Kuhn, S.; Horlacher, O.; Luttmann, E.; Willighagen, E. The chemistry development kit (CDK): An open-source Java library for chemo- and bioinformatics. J. Chem. Inf. Comput. Sci. 2003, 43, 493–500. [Google Scholar] [CrossRef]
- Steinbeck, C.; Hoppe, C.; Kuhn, S.; Floris, M.; Guha, R.; Willighagen, E.L. Recent developments of the Chemistry Development Kit (CDK)-an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006, 12, 2111–2120. [Google Scholar] [CrossRef]
- Willett, P.; Barnard, J.M.; Downs, G.M. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998, 38, 983–996. [Google Scholar] [CrossRef]
- Butina, D. Unsupervised data base clustering based on Daylight’s fingerprint and Tanimoto similarity: A fast and automated way to cluster small and large data sets. J. Chem. Inf. Comput. Sci. 1999, 39, 747–750. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Hildebrandt, C.; Wolf, S.; Neumann, S. Database supported candidate search for metabolite identification. J. Integr. Bioinforma. 2011, 8, 157. [Google Scholar] [CrossRef]
- Peironcely, J.E.; Rojas-Cherto, M.; Tas, A.; Vreeken, R.J.; Reijmers, T.; Coulier, L.; Hankemeier, T. An automated pipeline for de novo metabolite identification using mass spectrometry–based metabolomics. Anal. Chem. 2013, in press. [Google Scholar]
- RDCT. R Development Core Team; Institute for Statistics and Mathematics, Vienna University of Economics and Business: Vienna, Austria, 2013. Available online: http://www.r-project.org/ (accessed on 12 March 2013).
- Guha, R. Chemical informatics functionality in R. J. Stat. Softw. 2007, 18, 1–16. [Google Scholar]
- Schymanski, E.L.; Neumann, S. Critical Assessment of Small Molecule Identification Contest Rules. 2012. Available online: http://casmi-contest.org/rules.shtml (accessed on 20 May 2013).
- Allwood, J.W.; Weber, R.J.M.; Zhou, J.; He, S.; Viant, M.; Dunn, W.B. CASMI-the small molecule identification process from a Birmingham perspective. Metabolites 2013, 18, 397–411. [Google Scholar]
- Shen, H.; Zamboni, N.; Heinonen, M.; Rousu, J. Metabolite identification through machine learning-tackling CASMI challenges using FingerID. Metabolites 2013. submitted. [Google Scholar]
- Dührkop, K.; Scheubert, K.; Böcker, S. Molecular formula identification with SIRIUS. Metabolites 2013. submitted. [Google Scholar]
- Oberacher, H. Applying tandem mass spectral libraries for solving the CASMI LC/MS challenge 2012. Metabolites 2013, 3, 312–324. [Google Scholar] [CrossRef]
- Ruttkies, C.; Gerlich, M.; Neumann, S. Tackling challenging challenges with metfrag and metfusion. Metabolites 2013. submitted. [Google Scholar]
- Meringer, M.; Schymanski, E.L. Small molecule identification with MOLGEN and mass spectrometry. Metabolites 2013, in press. [Google Scholar]
- Schymanski, E.L.; Neumann, S. Critical Assessment of Small Molecule Identification Contest Results. 2013. Available online: http://casmi-contest.org/results.shtml/ (accessed on 20 May 2013).
- Brown, M.; Wedge, D.C.; Goodacre, R.; Kell, D.B.; Baker, P.N.; Kenny, L.C.; Mamas, M.A.; Neyses, L.; Dunn, W.B. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics 2011, 27, 1108–1112. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucl. Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Pence, H.E.; Williams, A. ChemSpider: An online chemical information resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Wolf, S.; Schmidt, S.; Müller-Hannemann, M.; Neumann, S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinform. 2010, 11, 148. [Google Scholar] [CrossRef]
- Heinonen, M.; Shen, H.; Zamboni, N.; Rousu, J. Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics 2012, 28, 2333–2341. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Böcker, S.; Letzel, M.C.; Lipták, Z.; Pervukhin, A. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics 2009, 25, 218–224. [Google Scholar] [CrossRef]
- Rasche, F.; Svatos, A.; Maddula, R.K.; Böttcher, C.; Böocker, S. Computing fragmentation trees from tandem mass spectrometry data. Anal. Chem. 2011, 83, 1243–1251. [Google Scholar] [CrossRef]
- NCBI. PubChem Compound Search; National Center for Biotechnology Information, U.S. National Library of Medicine: Bethesda, Maryland, USA, 2013. Available online: http://pubchem.ncbi.nlm.nih.gov/search/search.cgi# (accessed on 12 March 2013).
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- NIST/EPA/NIH. NIST 2011 Mass Spectral Library; National Institute of Standards and Technology, US Secretary of Commerce: Gaithersburg, Maryland, USA, 2011. [Google Scholar]
- Oberacher, H. Wiley Registry of Tandem Mass Spectral Data, MS for ID, 2012; ISBN: 978-1-1180-3744-7.
- Oberacher, H.; Pavlic, M.; Libiseller, K.; Schubert, B.; Sulyok, M.; Schuhmacher, R.; Csaszar, E.; Köfeler, H.C. On the inter-instrument and inter-laboratory transferability of a tandem mass spectral reference library: 1. Results of an Austrian multicenter study. J. Mass. Spectrom. 2009, 44, 485–493. [Google Scholar] [CrossRef]
- Meringer, M.; Reinker, S.; Zhang, J.; Muller, A. MS/MS data improves automated determination of molecular formulas by mass spectrometry. MATCH Commun. Math. Comput. Chem. 2011, 65, 259–290. [Google Scholar]
- Benecke, C.; Grüner, T.; Kerber, A.; Laue, R.; Wieland, T. MOLecular structure GENeration with MOLGEN, new features and future developments. Fresenius J. Anal. Chem. 1997, 359, 23–32. [Google Scholar] [CrossRef]
- Gugisch, R.; Kerber, A.; Kohnert, A.; Laue, R.; Meringer, M.; Rücker, C.; Wassermann, A. MOLGEN 5.0, a Molecular Structure Generator in Advances in Mathematical Chemistry. In Advances in Mathematical Chemistry; Basak, S.C., Restrepo, G., Villaveces, J.L., Eds.; Bentham Science Publishers: Sharjah, UAE, 2013; in press. [Google Scholar]
- Schymanski, E.; Gallampois, C.; Krauss, M.; Meringer, M.; Neumann, S.; Schulze, T.; Wolf, S.; Brack, W. Consensus structure elucidation combining GC/EI-MS, structure generation, and calculated properties. Anal. Chem. 2012, 84, 3287–3295. [Google Scholar] [CrossRef]
- Kerber, A.; Laue, R.; Meringer, M.; Rücker, C. MOLGEN–QSPR, a software package for the ucker, C. search of quantitative structure property relationships. Match Commun. Math. Comput. Chem. 2004, 51, 187–204. [Google Scholar]
- Kerber, A.; Laue, R.; Meringer, M.; Varmuza, K. MOLGEN–MS: Evaluation of low resolution electron impact mass spectra with ms classification and exhaustive structure generation. In Advances in Mass Spectrometry; Gelpi, E., Ed.; Wiley: West Sussex, UK, 2001; Volume 15, pp. 939–940. [Google Scholar]
- Schymanski, E.L.; Meinert, C.; Meringer, M.; Brack, W. The use of MS classifiers and structure generation to assist in the identification of unknowns in effect-directed analysis. Anal. Chim. Acta 2008, 615, 136–147. [Google Scholar] [CrossRef]
- USEPA. Estimation Program Interface (EPI) Suite v. 3.20; US Environmental Protection Agency: Washington, DC, USA, 2007.
- Peironcely, J.E.; Reijmers, T.; Coulier, L.; Bender, A.; Hankemeier, T. Understanding and classifying metabolite space and metabolite-likeness. PLoS One 2011, 6, e28966. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Schymanski, E.L.; Neumann, S. CASMI: And the Winner is . . . Metabolites 2013, 3, 412-439. https://doi.org/10.3390/metabo3020412
Schymanski EL, Neumann S. CASMI: And the Winner is . . . Metabolites. 2013; 3(2):412-439. https://doi.org/10.3390/metabo3020412
Chicago/Turabian StyleSchymanski, Emma L., and Steffen Neumann. 2013. "CASMI: And the Winner is . . ." Metabolites 3, no. 2: 412-439. https://doi.org/10.3390/metabo3020412