Glycerophosphoglycerol, Beta-Alanine, and Pantothenic Acid as Metabolic Companions of Glycolytic Activity and Cell Migration in Breast Cancer Cell Lines
Abstract
:1. Introduction
| Parameter | MCF-7 | MDA-MB-231 | MDA-MB-435 | MDA-MB-436 | JIMT-1 | Reference |
|---|---|---|---|---|---|---|
| Origin | PE | PE | PE | PE | PE | [2,21] |
| Tumor type | IDC | AC | IDC | IDC | AC | [2,21] |
| Differentiation grade | High | Poor | Poor | Poor | Poor | [2,21,22] |
| ER, PR, ERB/B2 status | ER+/PR+ | TN | TN | TN | ERB/B2+ | [23,24] |
| p53 status | wt | m | m | m | m | [2,23] |
| In vitro invasiveness | +/ ++ | +++/ +++++ | +++/ +++ | ++/ +++ | n.a. n.a. | [22] [24] |
| Tumors in nude mice | P | LI | LI | LI | Yes, not further specified | [21,24] |
| Gene cluster | luminal | basal B | basal B | basal B | basal A/ ERB/B2 | [2,25] |
2. Experimental Section
2.1. Materials and Chemicals
2.2. Cell Culture
2.3. Stable Transfection of MCF-7 Cells with a shRNA-Expressing Plasmid for GLO1 Silencing
2.4. Metabolite Extraction, Derivatization and GCMS Analysis
2.5. Determination of Medium pH
2.6. Protein Extraction, Determination of Protein Content and Immunoblotting
2.7. Enzyme Activity Measurements
2.8. RNA Isolation, Reverse Transcription, Semi-Quantitative and qRT-PCR
2.9. Migration Assay
2.10. Sample Evaluation and Statistical Analysis
3. Results and Discussion
3.1. Metabolite Profiles of Cancer Cell Lines are Cell-Type Specific

| Cell line | Analyte | Relative abundance | p-values sample set 1 | p-values sample set 2 |
|---|---|---|---|---|
| MCF-7 | Cystathione (2TMS) | Increased | n.d. | 1.2×10−6 |
| Asparagine DL (3TMS) | Increased | 0.05 | 2.6×10−4 | |
| Gulonic acid (6TMS) | Increased | 0.37 | 1.3×10−3 | |
| Piperidine-2-one, 3-amino (2TMS) | Increased | n.d. | 6.6×10−3 | |
| Proline 4-hydroxy-(3TMS) | Increased | 0.03 | 5.8×10−5 | |
| Hexadecan-1-ol, n- (1TMS) | Increased | 0.02 | 4.4×10−4 | |
| Arginine DL --NH3 (3TMS) | Increased | n.d. | 2.6×10−3 | |
| Ornithine DL (4TMS) | Increased | 0.19 | 6.6×10−5 | |
| Ornithine DL (3TMS) | Increased | n.d. | 5.0×10−3 | |
| Inositol myo (6TMS) | Decreased | 6.2×10-13 | 6.6×10−8 | |
| Alanine beta (3TMS) | Decreased | 2.6×10-5 | 1.7×10−9 | |
| Octadecan-1-ol, n- (1TMS) | Increased | 0.04 | 2.4×10−4 | |
| JIMT-1 | Pantothenic acid D (3TMS) | Increased | 0.08 | 1.2×10−5 |
| Alanine (2TMS) | Increased | 1.4×10-4 | 5.3×10−4 | |
| Glutaric acid, 2-hydroxy (3TMS) | Increased | n.d. | 2.1×10−9 | |
| Glutamine DL (4TMS) | Decreased | n.d. | 9.3×10−6 |
3.2. Lactate Release Is Enhanced for MDA-MB-231 and JIMT-1
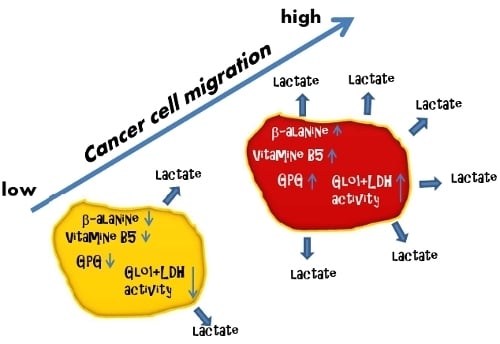
3.4. Cell Migration Correlates with Extracellular Lactate Concentrations, LDH and Glo1 Activity, and Glycerophosphoglycerol, β-Alanine and Pantothenic Acid
| Sample set | extracellular lactate* | Glo1 activity | LDH activity | Cell migration | |||||
|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | ||
| Glycerophosphoglycerol (5TMS) | 2 | 0.77 | 0.009 | 0.87 | 0.001 | 0.91 | 9.50×10−5 | 0.98 | 5.03×10−7 |
| 1 | 0.81 | 0.085 | 0.88 | 0.016 | 0.95 | 0.002 | 0.78 | 0.057 | |
| Pantothenic acid D (3TMS) | 2 | 0.87 | 0.001 | 0.84 | 0.001 | 0.85 | 0.001 | 0.86 | 0.001 |
| 1 | 0.67 | 0.282 | 0.91 | 0.007 | 0.85 | 0.026 | 0.95 | 0.002 | |
| Alanine, beta- (3TMS) | 2 | 0.91 | 1.24×10−4 | 0.63 | 0.069 | 0.82 | 0.003 | 0.82 | 0.003 |
| 1 | 0.99 | 2.38×10−4 | 0.63 | 0.204 | 0.85 | 0.023 | 0.63 | 0.206 | |
| extracellular lactate | 2 | 0.84 | 0.002 | 0.80 | 0.004 | 0.85 | 0.001 | ||
| 1 | 0.72 | 0.104 | 0.89 | 0.013 | 0.82 | 0.037 | |||
| Glo1 activity | 2 | 0.84 | 0.002 | 0.76 | 0.010 | 0.86 | 0.001 | ||
| 1 | 0.72 | 0.197 | 0.94 | 0.004 | 0.86 | 0.020 | |||
| LDH activity | 2 | 0.80 | 0.004 | 0.76 | 0.010 | 0.92 | 9.25×10−5 | ||
| 1 | 0.89 | 0.029 | 0.94 | 0.004 | 0.81 | 0.040 | |||
| Cell migration | 2 | 0.85 | 0.001 | 0.86 | 0.001 | 0.92 | 9.25×10−5 | ||
| 1 | 0.82 | 0.078 | 0.86 | 0.020 | 0.81 | 0.040 | |||
4. Conclusions
Acknowledgments
Conflicts of Interest
References and Notes
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.-P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumors. Nature 2000, 406, 747–752. [Google Scholar]
- Sims, A.H.; Howell, A.; Howell, S.J.; Clarke, R.B. Origins of breast cancer subtypes and therapeutic implications. Nat. Clin. Pract. Oncol. 2007, 4, 516–525. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar]
- Yang, C.; Richardson, A.D.; Smith, J.W.; Osterman, A. Comparative metabolomics of breast cancer. Pac. Symp. Biocomput. 2007, 12, 181–192. [Google Scholar]
- Engel, N.; Lisec, J.; Piechulla, B.; Nebe, B. Metabolic Profiling Reveals Sphingosine-1-Phosphate Kinase 2 and Lyase as Key Targets of (Phyto-) Estrogen Action in the Breast Cancer Cell Line MCF-7 and Not in MCF-12A. PLoS One 2012, 7, e47833. [Google Scholar]
- Cao, M.D.; Döpkens, M.; Krishnamachary, B.; Vesuna, F.; Gadiya, M.M.; Lønning, P.E.; Bhujwalla, Z.M.; Gribbestad, I.S.; Glunde, K. Glycerophosphodiester phosphodiesterase domain containing 5 (GDPD5) expression correlates with malignant choline phospholipid metabolite profiles in human breast cancer. NMR Biomed. 2012, 25, 1033–1042. [Google Scholar] [Green Version]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef]
- Griffin, J.L.; Shockcor, J.P. Metabolic profiles of cancer cells. Nat. Rev. Canc. 2004, 4, 551–561. [Google Scholar] [CrossRef]
- Asiago, V.M.; Alvarado, L.Z.; Shanaiah, N.; Gowda, G.A.N.; Owusu-Sarfo, K.; Ballas, R.A.; Raftery, D. Early Detection of Recurrent Breast Cancer Using Metabolite Profiling. Cancer Res. 2010, 70, 8309–8318. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar]
- Isidoro, A.; Casado, E.; Redondo, A.; Acebo, P.; Espinosa, E.; Alonso, A.M.; Cejas, P.; Hardisson, D.; Fresno Vara, J.A.; Belda-Iniesta, C.; et al. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis 2005, 26, 2095–2104. [Google Scholar] [CrossRef]
- Thornalley, P.J.; Rabbani, N. Glyoxalase in tumorigenesis and multidrug resistance. Semin. Cell. Dev. Biol. 2011, 22, 318–325. [Google Scholar] [CrossRef]
- Bair, W.B.I.; Cabello, C.M.; Uchida, K.; Bause, A.S.; Wondrak, G.T. GLO1 overexpression in human malignant melanoma. Melanoma Res. 2010, 20, 85–96. [Google Scholar] [CrossRef]
- Jones, M.B.; Krutzsch, H.; Shu, H.; Zhao, Y.; Liotta, L.A.; Kohn, E.C.; Petricoin, E.F. Proteomic analysis and identification of new biomarkers and therapeutic targets for invasive ovarian cancer. Proteomics 2002, 2, 76–84. [Google Scholar]
- Sakamoto, H.; Mashima, T.; Sato, S.; Hashimoto, Y.; Yamori, T.; Tsuruo, T. Selective Activation of Apoptosis Program by S-p-bromobenzylglutathione Cyclopentyl Diester in Glyoxalase I-overexpressing Human Lung Cancer Cells. Clin. Cancer Res. 2001, 7, 2513–2518. [Google Scholar]
- Cheng, W.-L.; Tsai, M.-M.; Tsai, C.-Y.; Huang, Y.-H.; Chen, C.-Y.; Chi, H.-C.; Tseng, Y.-H.; Chao, I.-W.; Lin, W.-C.; Wu, S.-M.; et al. Glyoxalase-I Is a Novel Prognosis Factor Associated with Gastric Cancer Progression. PLoS One 2012, 7, e34352. [Google Scholar] [CrossRef]
- Arseneault, R.; Chien, A.; Newington, J.T.; Rappon, T.; Harris, R.; Cumming, R.C. Attenuation of LDHA expression in cancer cells leads to redox-dependent alterations in cytoskeletal structure and cell migration. Cancer Lett. 2013, 338, 255–266. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Tanner, M.; Kapanen, A.I.; Junttila, T.; Raheem, O.; Grenman, S.; Elo, J.; Elenius, K.; Isola, J. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol. Cancer Ther. 2004, 3, 1585–1592. [Google Scholar]
- Sommers, C.L.; Byers, S.W.; Thompson, E.W.; Torri, J.A.; Gelmann, E.R. Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res. Tr. 1994, 31, 325–335. [Google Scholar]
- Hu, X.; Stern, H.M.; Ge, L.; O’Brien, C.; Haydu, L.; Honchell, C.D.; Haverty, P.M.; Peters, B.A.; Wu, T.D.; Amler, L.C.; et al. Genetic Alterations and Oncogenic Pathways Associated with Breast Cancer Subtypes. Mol. Cancer Res. 2009, 7, 511–522. [Google Scholar] [CrossRef]
- Thompson, E.W.; Paik, S.; Brunner, N.; Sommers, C.L.; Zugmaier, G.; Clarke, R.; Shima, T.B.; Torri, J.; Donahue, S.; Lippman, M.E.; et al. Association of increased basement membrane invasiveness with abscence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J. Cell. Physiol. 1992, 150, 534–544. [Google Scholar] [CrossRef]
- Olsson, E.; Honeth, G.; Bendahl, P.-O.; Saal, L.; Gruvberger-Saal, S.; Ringner, M.; Vallon-Christersson, J.; Jonsson, G.; Holm, K.; Lovgren, K.; et al. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer 2011, 11, 418. [Google Scholar] [CrossRef]
- Aboagye, E.O.; Bhujwalla, Z.M. Malignant Transformation Alters Membrane Choline Phospholipid Metabolism of Human Mammary Epithelial Cells. Cancer Res. 1999, 59, 80–84. [Google Scholar]
- Dolce, V.; Cappello, A.R.; Lappano, R.; Maggiolini, M. Glycerophospholipid synthesis as a novel drug target against cancer. Curr. Mol. Pharmacol. 2011, 4, 167–175. [Google Scholar]
- Mashima, T.; Seimiya, H.; Tsuruo, T. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br. J. Cancer 2009, 100, 1369–1372. [Google Scholar] [CrossRef]
- Hutschenreuther, A.; Kiontke, A.; Birkenmeier, G.; Birkemeyer, C. Comparison of extraction conditions and normalization approaches for cellular metabolomics of adherent growing cells with GC-MS. Anal. Methods 2012, 4, 1953–1963. [Google Scholar]
- Van de Wetering, M.; Oving, I.; Muncan, V.; Pon Fong, M.T.; Brantjes, H.; van Leenen, D.; Holstege, F.C.P.; Brummelkamp, T.R.; Agami, R.; Clevers, H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003, 4, 609–615. [Google Scholar] [CrossRef]
- Santel, T.; Pflug, G.; Hemdan, N.Y.A.; Schäfer, A.; Hollenbach, M.; Buchold, M.; Hintersdorf, A.; Lindner, I.; Otto, A.; Bigl, M.; et al. Curcumin Inhibits Glyoxalase 1—A Possible Link to Its Anti-Inflammatory and Anti-Tumor Activity. PLoS One 2008, 3, e3508. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mannervik, B.; Aronsson, A.-C.; Tibbelin, G. Glyoxalase I from Human Erythrocytes. Method. Enzymol. 1982, 90, 535–541. [Google Scholar] [CrossRef]
- Lindner, I.; Hemdan, N.Y.A.; Buchold, M.; Huse, K.; Bigl, M.; Oerlecke, I.; Ricken, A.; Gaunitz, F.; Sack, U.; Naumann, A.; et al. α2-Macroglobulin Inhibits the Malignant Properties of Astrocytoma Cells by Impeding β-Catenin Signaling. Cancer Res. 2010, 70, 277–287. [Google Scholar] [CrossRef]
- SDS Software, version 1.4, software of Applied Biosystems 7500/7500 Fast Real-Time PCR. Applied Biosystems: Foster City, CA, USA, 2006.
- IBM SPSS Statistics, version 19, IBM: Chicago, IL, USA, 2010.
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Phang, J.M.; Liu, W.; Zabirnyk, O. Proline Metabolism and Microenvironmental Stress. Annu. Rev. Nutr. 2010, 30, 441–463. [Google Scholar] [CrossRef]
- Richardson, A.D.; Yang, C.; Osterman, A.; Smith, J.W. Central carbon metabolism in the progression of mammary carcinoma. Breast Cancer Res. Tr. 2008, 110, 297–307. [Google Scholar] [CrossRef]
- Shapiro, I.M.; Cheng, A.W.; Flytzanis, N.C.; Balsamo, M.; Condeelis, J.S.; Oktay, M.H.; Burge, C.B.; Gertler, F.B. An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype. PLoS Genet. 2011, 7, e1002218. [Google Scholar] [CrossRef] [Green Version]
- Hori, S.; Nishiumi, S.; Kobayashi, K.; Shinohara, M.; Hatakeyama, Y.; Kotani, Y.; Hatano, N.; Maniwa, Y.; Nishio, W.; Bamba, T.; et al. A metabolomic approach to lung cancer. Lung Cancer 2011, 74, 284–292. [Google Scholar] [CrossRef]
- Kao, J.; Salari, K.; Bocanegra, M.; Choi, Y.-L.; Girard, L.; Gandhi, J.; Kwei, K.A.; Hernandez-Boussard, T.; Wang, P.; Gazdar, A.F.; et al. Molecular Profiling of Breast Cancer Cell Lines Defines Relevant Tumor Models and Provides a Resource for Cancer Gene Discovery. PLoS One 2009, 4, e6146. [Google Scholar] [CrossRef]
- Feron, O. Pyruvate into lactate and back: From the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiother. Oncol. 2009, 92, 329–333. [Google Scholar] [CrossRef]
- Santarius, T.; Bignell, G.R.; Greenman, C.D.; Widaa, S.; Chen, L.; Mahoney, C.L.; Butler, A.; Edkins, S.; Waris, S.; Thornalley, P.J.; et al. GLO1—A novel amplified gene in human cancer. Gene. Chromosome. Canc. 2010, 49, 711–725. [Google Scholar] [CrossRef]
- Rulli, A.; Carli, L.; Romani, R.; Baroni, T.; Giovannini, E.; Rosi, G.; Talesa, V. Expression of glyoxalase I and II in normal and breast cancer tissues. Breast Cancer Res. Tr. 2001, 66, 67–72. [Google Scholar] [CrossRef]
- Birkenmeier, G.; Stegemann, C.; Hoffmann, R.; Günther, R.; Huse, K.; Birkemeyer, C. Posttranslational Modification of Human Glyoxalase 1 Indicates Redox-Dependent Regulation. PLoS One 2010, 5, e10399. [Google Scholar]
- Parra-Bonilla, G.; Alvarez, D.F.; Al-Mehdi, A.-B.; Alexeyev, M.; Stevens, T. Critical role for lactate dehydrogenase A in aerobic glycolysis that sustains pulmonary microvascular endothelial cell proliferation. Am. J. Physiol. Lung Cell Mol. Physiol. 2010, 299, L513–L522. [Google Scholar]
- Semenza, G.L. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 2007, 405, 1–9. [Google Scholar]
- Ayoub, F.; Zaman, M.; Thornalley, P.; Masters, J. Glyoxalase activities in human tumor cell lines in vitro. Anticancer Res. 1993, 13, 151–155. [Google Scholar]
- Kim, J.-W.; Dang, C.V. Cancer's Molecular Sweet Tooth and the Warburg Effect. Cancer Res. 2006, 66, 8927–8930. [Google Scholar] [CrossRef]
- Beckner, M.E.; Stracke, M.L.; Liotta, L.A.; Schiffmann, E. Glycolysis as Primary Energy Source in Tumor Cell Chemotaxis. J. Natl. Cancer Inst. 1990, 82, 1836–1840. [Google Scholar] [CrossRef]
- Zhang, D.; Tai, L.K.; Wong, L.L.; Chiu, L.-L.; Sethi, S.K.; Koay, E.S.C. Proteomic Study Reveals That Proteins Involved in Metabolic and Detoxification Pathways Are Highly Expressed in HER-2/neu-positive Breast Cancer. Mol. Cell. Proteomics 2005, 4, 1686–1696. [Google Scholar] [CrossRef]
- Slyshenkov, V.S.; Omelyanchik, S.N.; Moiseenok, A.G.; Trebukhina, R.V.; Wojtczak, L. Pantothenol Protects Rats Against Some Deleterious Effects of Gamma Radiation. Free Radic. Biol. Med. 1998, 24, 894–899. [Google Scholar] [CrossRef]
- Slyshenkov, V.S.; Rakowska, M.; Moiseenok, A.G.; Wojtczak, L. Pantothenic acid and its derivatives protect Ehrlich ascites tumor cells against lipid peroxidation. Free Radic. Biol. Med. 1995, 19, 767–772. [Google Scholar] [CrossRef]
- Slyshenkov, V.S.; Moiseenok, A.G.; Wojtczak, L. Noxious effects of oxygen reactive species on energy-coupling processes in Ehrlich ascites tumor mitochondria and the protection by pantothenic acid. Free Radic. Biol. Med. 1996, 20, 793–800. [Google Scholar] [CrossRef]
- Sliva, D.; Mason, R.; Xiao, H.; English, D. Enhancement of the Migration of Metastatic Human Breast Cancer Cells by Phosphatidic Acid. Biochem. Biophys. Res. Comm. 2000, 268, 471–479. [Google Scholar] [CrossRef]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hutschenreuther, A.; Birkenmeier, G.; Bigl, M.; Krohn, K.; Birkemeyer, C. Glycerophosphoglycerol, Beta-Alanine, and Pantothenic Acid as Metabolic Companions of Glycolytic Activity and Cell Migration in Breast Cancer Cell Lines. Metabolites 2013, 3, 1084-1101. https://doi.org/10.3390/metabo3041084
Hutschenreuther A, Birkenmeier G, Bigl M, Krohn K, Birkemeyer C. Glycerophosphoglycerol, Beta-Alanine, and Pantothenic Acid as Metabolic Companions of Glycolytic Activity and Cell Migration in Breast Cancer Cell Lines. Metabolites. 2013; 3(4):1084-1101. https://doi.org/10.3390/metabo3041084
Chicago/Turabian StyleHutschenreuther, Antje, Gerd Birkenmeier, Marina Bigl, Knut Krohn, and Claudia Birkemeyer. 2013. "Glycerophosphoglycerol, Beta-Alanine, and Pantothenic Acid as Metabolic Companions of Glycolytic Activity and Cell Migration in Breast Cancer Cell Lines" Metabolites 3, no. 4: 1084-1101. https://doi.org/10.3390/metabo3041084