Carbon Partitioning in Green Algae (Chlorophyta) and the Enolase Enzyme
Abstract
:1. Introduction

2. Results and Discussion
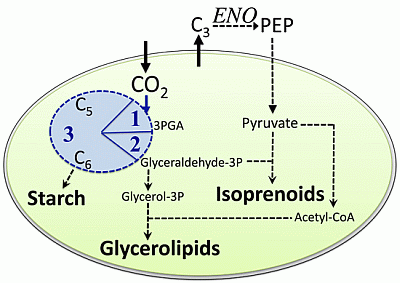
2.1. Carbon Partitioning in Green Algae

2.2. The Enolase
- (1)
- A plastid-localized enolase (ENO1) [53].
- (2)
- (3)
- An enolase localized to the cytosol (ENOc [16]),
- (4)
- A multi-functional enolase (DEP1) [55], which was detected in the chloroplast stroma in A. thaliana [55]. DEP1 in Arabidopsis functions in the methionine biosynthesis [56] performing the reaction of the 5’-methylthioribulose-1-phosphate dehydratase (EC 4.2.1.109) and of the Enolase-phosphatase E1 (EC 3.1.3.77). The DEP1 enolase activity converts 2,3-diketo-5-methylthiopentyl-1-phosphate into the intermediate 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate, which is then dephosphorylated. DEP1 is an essential enzyme in cellular sulfur metabolism in plants [57].
2.3. The Enolase in Green Algae
| Species | JGI Protein ID | JGI Locus Name & Location | Notes |
|---|---|---|---|
| Asterochloris sp. v1.0 | 35232 | scaffold_00088:18492-25512 [JGI Genome Portal] | Glycolytic enolase |
| Chlamydomonas reinhardtii | 136652 | Cre12.g513200.t1.2; chromosome_12: 3521433-3525524 [Phytozome v9.1] | Glycolytic enolase |
| Chlamydomonas reinhardtii | NA | g393; chromosome_1: 2781663–2787514 [Phytozome v9.1] | Multi-functional enolase |
| Chlorella variabilis NC64A | 136652 | scaffold_17:245647-250804 [JGI Genome Portal] | Glycolytic enolase; Gaps in genomic sequence |
| Coccomyxa subelipsoidea | 38308 | fgenesh1_pm.19_#_130; scaffold_19: 1328456–1334257 [Phytozome v9.1] | Glycolytic enolase |
| Coccomyxa subelipsoidea C-169 | 35576 | fgenesh1_pm.3_#_251; scaffold_3: 2788580–2794381 [Phytozome v9.1] | Glycolytic enolase |
| Coccomyxa subelipsoidea C-169 | NA | estExt_Genewise1Plus.C_20589; scaffold_2: 2932335–2935743 [Phytozome v9.1] | Multi-functional enolase |
| Micromonas pusilla CCMP1545 | 122580 | scaffold_2: 722515–724505 [Phytozome v9.1] | Glycolytic enolase |
| Micromonas pusilla RCC299 | 107587 | Chr_01: 1470509–1472948 [Phytozome v9.1] | Glycolytic enolase |
| Ostreococcus lucimarinus | 28765 | Chr_1: 271278–273263 [Phytozome v9.1] | Glycolytic enolase |
| Ostreococcus tauri v2.0 | 27349 | Chr_01.0001:228132-230031 [JGI Genome Portal] | Glycolytic enolase |
| Volvox carteri | 79991 | Vocar20013958m.g; scaffold_6: 3504767–3509879 [Phytozome v9.1] | Glycolytic enolase |
| Volvox carteri | 42159 | Vocar20006493m.g; scaffold_7: 3486914–3491916 [Phytozome v9.1] | Multi-functional enolase |
| Dunaliella salina CCAP19/18 | - | GenBank accession number KM008612 | Glycolytic enolase |
| Dunaliella salina CCAP19/18 | - | GenBank accession number KM008613 | Multi-functional enolase |



3. Experimental Section
3.1. Comparative Genomics
3.2. Phylogenetic Analysis
3.3. Structural Modeling of the Enolase Enzyme
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Georgianna, D.R.; Mayfield, S.P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 2012, 488, 329–335. [Google Scholar] [CrossRef]
- Klein-Marcuschamer, D. A matter of detail: Assessing the true potential of microalgal biofuels. Biotechnol. Bioeng. 2013, 110, 2317–2318. [Google Scholar] [CrossRef]
- Stephens, E.; Ross, I.L.; Hankamer, B. Expanding the microalgal industry-continuing controversy or compelling case? Curr. Opin. Chem. Biol. 2013, 17, 444–452. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Symbiogenesis: Mechanisms, evolutionary consequences, and systematic implications. In Annual Review of Ecology and Systematics; Futuyma, D.J., Ed.; Annual Reviews: Palo Alto, CA, USA, 2013; Volume 44, p. 145. [Google Scholar]
- Keeling, P.J. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Annual Reviews: Palo Alto, CA, USA, 2013; Volume 64, pp. 583–607. [Google Scholar]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; de Clerck, O. Phylogeny and molecular evolution of the green algae. CRC Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Lohr, M.; Schwender, J.; Polle, J.E.W. Isoprenoid biosynthesis in eukaryotic phototrophs: A spotlight on algae. Plant Sci. 2012, 185, 9–22. [Google Scholar]
- Milo, R.; Last, R.L. Achieving diversity in the face of constraints: Lessons from metabolism. Science 2012, 336, 1663–1667. [Google Scholar] [CrossRef]
- Becker, E.W. Chemical composition. In Microalgae: Biotechnology and Microbiology; Cambridge University Press: Cambridge, England, 1994; Chapter 12; pp. 177–195. [Google Scholar]
- Sheehan, J.; Dunahay, T.; Benemann, J.; Roessler, P. A Look Back at the US Department of Energy’s Aquatic Species Program-Biodiesel from Algae; National Renewable Energy Laboratory: Golden, USA, 1998. [Google Scholar]
- Guerra, L.T.; Levitan, O.; Frada, M.J.; Sun, J.S.; Falkowski, P.G.; Dismukes, G.C. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy 2013, 59, 306–315. [Google Scholar] [CrossRef]
- Li, Y.T.; Han, D.X.; Sommerfeld, M.; Hu, Q.A. Photosynthetic carbon partitioning and lipid production in the oleaginous microalga Pseudochlorococcum sp. (Chlorophyceae) under nitrogen-limited conditions. Bioresour. Technol. 2011, 102, 123–129. [Google Scholar] [CrossRef]
- Packer, A.; Li, Y.T.; Andersen, T.; Hu, Q.A.; Kuang, Y.; Sommerfeld, M. Growth and neutral lipid synthesis in green microalgae: A mathematical model. Bioresour. Technol. 2011, 102, 111–117. [Google Scholar] [CrossRef]
- Melis, A. Carbon partitioning in photosynthesis. Curr. Opin. Chem. Biol. 2013, 17, 453–456. [Google Scholar] [CrossRef]
- Prabhakar, V.; Lottgert, T.; Geimer, S.; Dormann, P.; Kruger, S.; Vijayakumar, V.; Schreiber, L.; Gobel, C.; Feussner, K.; Feussner, I.; et al. Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell 2010, 22, 2594–2617. [Google Scholar] [CrossRef]
- Voll, L.M.; Hajirezaei, M.R.; Czogalla-Peter, C.; Lein, W.; Stitt, M.; Sonnewald, U.; Bornke, F. Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytol. 2009, 184, 607–618. [Google Scholar] [CrossRef]
- O’Grady, J.; Schwender, J.; Shachar-Hill, Y.; Morgan, J.A. Metabolic cartography: Experimental quantification of metabolic fluxes from isotopic labelling studies. J. Exp. Bot. 2012, 63, 2293–2308. [Google Scholar] [CrossRef]
- Winck, F.V.; Melo, D.O.P.; Barrios, A.F.G. Carbon acquisition and accumulation in microalgae Chlamydomonas: Insights from “omics” approaches. J. Proteomics 2013, 94, 207–218. [Google Scholar] [CrossRef]
- Dal’Molin, C.G.D.; Quek, L.E.; Palfreyman, R.W.; Nielsen, L.K. Algagem-a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome. BMC Genomics 2011. [Google Scholar] [CrossRef]
- Johnson, X.; Alric, J. Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: Metabolic constraints for carbon partitioning between oil and starch. Eukaryotic Cell 2013, 12, 776–793. [Google Scholar] [CrossRef]
- Kliphuis, A.M.J.; Klok, A.J.; Martens, D.E.; Lamers, P.P.; Janssen, M.; Wijffels, R.H. Metabolic modeling of Chlamydomonas reinhardtii: Energy requirements for photoautotrophic growth and maintenance. J. Appl. Phycol. 2012, 24, 253–266. [Google Scholar] [CrossRef]
- Christian, N.; May, P.; Kempa, S.; Handorf, T.; Ebenhoh, O. An integrative approach towards completing genome-scale metabolic networks. Mol. Biosyst. 2009, 5, 1889–1903. [Google Scholar] [CrossRef]
- Wienkoop, S.; Weiss, J.; May, P.; Kempa, S.; Irgang, S.; Recuenco-Munoz, L.; Pietzke, M.; Schwemmer, T.; Rupprecht, J.; Egelhofer, V.; et al. Targeted proteomics for Chlamydomonas reinhardtii combined with rapid subcellular protein fractionation, metabolomics and metabolic flux analyses. Mol. Biosyst. 2010, 6, 1018–1031. [Google Scholar] [CrossRef]
- Veyel, D.; Erban, A.; Fehrle, I.; Kopka, J.; Schroda, M. Rationale approaches for studying metabolism in eukaryotic microalgae. Metabolites 2014, 4, 184–217. [Google Scholar] [CrossRef]
- Pazour, G.J.; Agrin, N.; Leszyk, J.; Witman, G.B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005, 170, 103–113. [Google Scholar] [CrossRef]
- Terashima, M.; Specht, M.; Hippler, M. The chloroplast proteome: A survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr. Genet. 2011, 57, 151–168. [Google Scholar] [CrossRef]
- Klein, U. Compartmentation of glycolysis and of the oxidative pentose-phosphate pathway in Chlamydomonas reinhardii. Planta 1986, 167, 81–86. [Google Scholar] [CrossRef]
- Klock, G.; Kreuzberg, K. Compartmented metabolite pools in protoplasts from the green-alga Chlamydomonas reinhardtii-changes after transition from aerobiosis to anaerobiosis in the dark. Biochim. Biophys. Acta 1991, 1073, 410–415. [Google Scholar] [CrossRef]
- Mitchell, B.F.; Pedersen, L.B.; Feely, M.; Rosenbaum, J.L.; Mitchell, D.R. ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell 2005, 16, 4509–4518. [Google Scholar] [CrossRef]
- Ben-Amotz, A.; Katz, A.; Avron, M. Accumulation of beta-carotene in halotolerant algae-purification and characterization of beta-carotene-rich globules from Dunaliella bardawil (Chlorophyceae). J. Phycol. 1982, 18, 529–537. [Google Scholar] [CrossRef]
- Diaz-Ramos, A.; Roig-Borrellas, A.; Garcia-Melero, A.; Lopez-Alemany, R. Alpha-enolase, a multifunctional protein: Its role on pathophysiological situations. J. Biomed. Biotechnol. 2012. [Google Scholar] [CrossRef]
- Gerlt, J.A.; Allen, K.N.; Almo, S.C.; Armstrong, R.N.; Babbitt, P.C.; Cronan, J.E.; Dunaway-Mariano, D.; Imker, H.J.; Jacobson, M.P.; Minor, W.; et al. The enzyme function initiative. Biochemistry 2011, 50, 9950–9962. [Google Scholar] [CrossRef]
- Huberts, D.; van der Klei, I.J. Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta-Mol. Cell Res. 2010, 1083, 520–525. [Google Scholar] [CrossRef]
- Henderson, B.; Martin, A. Bacterial moonlighting proteins and bacterial virulence. In Between Pathogenicity and Commensalism; Dobrindt, U., Hacker, J.H., Svanborg, C., Eds.; 2013; Volume 358, pp. 155–213. [Google Scholar]
- Gomez-Arreaza, A.; Acosta, H.; Quinones, W.; Concepcion, J.L.; Michels, P.A.M.; Avilan, L. Extracellular functions of glycolytic enzymes of parasites: Unpredicted use of ancient proteins. Mol. Biochem. Parasitol. 2014, 193, 75–81. [Google Scholar] [CrossRef]
- Carneiro, C.R.W.; Postol, E.; Nomizo, R.; Reis, L.F.L.; Brentani, R.R. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 2004, 6, 604–608. [Google Scholar] [CrossRef]
- Ehinger, S.; Schubert, W.D.; Bergmann, S.; Hammerschmidt, S.; Heinz, D.W. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: Crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 2004, 343, 997–1005. [Google Scholar] [CrossRef]
- Pancholi, V.; Fischetti, V.A. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic Streptococci. J. Biol. Chem. 1998, 273, 14503–14515. [Google Scholar] [CrossRef]
- Swenerton, R.K.; Zhang, S.Y.; Sajid, M.; Medzihradszky, K.F.; Craik, C.S.; Kelly, B.L.; McKerrow, J.H. The oligopeptidase b of Leishmania regulates parasite enolase and immune evasion. J. Biol. Chem. 2011, 286, 429–440. [Google Scholar] [CrossRef]
- Vanegas, G.; Quinones, W.; Carrasco-Lopez, C.; Concepcion, J.L.; Albericio, F.; Avilan, L. Enolase as a plasminogen binding protein in Leishmania mexicana. Parasitol. Res. 2007, 101, 1511–1516. [Google Scholar] [CrossRef]
- Castillo-Romero, A.; Davids, B.J.; Lauwaet, T.; Gillin, F.D. Importance of enolase in Giardia lamblia differentiation. Mol. Biochem. Parasitol. 2012, 184, 122–125. [Google Scholar]
- Mathur, R.L.; Reddy, M.C.; Yee, S.Y.; Imbesi, R.; Grothvasselli, B.; Farnsworth, P.N. Investigation of lens glycolytic-enzymes-species distribution and interaction with supramolecular order. Exp. Eye Res. 1992, 54, 253–260. [Google Scholar] [CrossRef]
- Subramanian, A.; Miller, D.M. Structural analysis of alpha-enolase-mapping the functional domains involved in down-regulation of the c-myc protooncogene. J. Biol. Chem. 2000, 275, 5958–5965. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Steele, R.; Ray, R.B. Functional domains of c-myc promoter binding protein 1 involved in transcriptional repression and cell growth regulation. Mol. Cell. Biol. 1999, 19, 2880–2886. [Google Scholar]
- Kang, M.Y.; Abdelmageed, H.; Lee, S.; Reichert, A.; Mysore, K.S.; Allen, R.D. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 2013, 76, 481–493. [Google Scholar]
- Wichelecki, D.J.; Balthazor, B.M.; Chau, A.C.; Vetting, M.W.; Fedorov, A.A.; Fedorov, E.V.; Lukk, T.; Patskovsky, Y.V.; Stead, M.B.; Hillerich, B.S.; et al. Discovery of function in the enolase superfamily: D-mannonate and d-gluconate dehydratases in the d-mannonate dehydratase subgroup. Biochemistry 2014, 53, 2722–2731. [Google Scholar] [CrossRef]
- Wold, F. Enolase. Enzymes 1971, 5, 499–538. [Google Scholar]
- Pancholi, V. Multifunctional alpha-enolase: Its role in diseases. Cell. Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef]
- Chai, G.Q.; Brewer, J.M.; Lovelace, L.L.; Aoki, T.; Minor, W.; Lebioda, L. Expression, purification and the 1.8 angstrom resolution crystal structure of human neuron specific enolase. J. Mol. Biol. 2004, 341, 1015–1021. [Google Scholar]
- Ruan, K.; Duan, J.B.; Bai, F.; Lemaire, M.; Ma, X.; Bai, L. Function of Dunaliella salina (Dunaliellaceae) enolase and its expression during stress. Eur. J. Phycol. 2009, 44, 207–214. [Google Scholar] [CrossRef]
- TAIR. Available online: http://www.arabidopsis.org (accessed on 31 July 2014).
- Andriotis, V.M.E.; Kruger, N.J.; Pike, M.J.; Smith, A.M. Plastidial glycolysis in developing Arabidopsis embryos. New Phytol. 2010, 185, 649–662. [Google Scholar] [CrossRef]
- Prabhakar, V.; Lottgert, T.; Gigolashvili, T.; Bell, K.; Flugge, U.-I.; Hausler, R.E. Molecular and functional characterization of the plastid-localized Phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Letters 2009, 583, 983–991. [Google Scholar] [CrossRef]
- Vanderstraeten, D.; Rodriguespousada, R.A.; Goodman, H.M.; Vanmontagu, M. Plant enolase-gene structure, expression, and evolution. Plant Cell 1991, 3, 719–735. [Google Scholar] [CrossRef]
- Rutschow, H.; Ytterberg, A.J.; Friso, G.; Nilsson, R.; van Wijk, K.J. Quantitative proteomics of a chloroplast SRP54 sorting mutant and its genetic interactions with clpc1 in Arabidopsis. Plant Physiol. 2008, 148, 156–175. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Feussner, K.; Zierer, W.; Rabinovych, V.; Klebl, F.; Feussner, I.; Sauer, N. Phloem-specific expression of yang cycle genes and identification of novel yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 2011, 23, 1904–1919. [Google Scholar] [CrossRef]
- Sauter, M.; Moffatt, B.; Saechao, M.C.; Hell, R.; Wirtz, M. Methionine salvage and s-adenosylmethionine: Essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013, 451, 145–154. [Google Scholar]
- Troncoso-Ponce, M.A.; Garces, R.; Martinez-Force, E. Glycolytic enzymatic activities in developing seeds involved in the differences between standard and low oil content sunflowers (Helianthus annuus L.). Plant Physiol. Biochem. 2010, 48, 961–965. [Google Scholar] [CrossRef]
- Harper, J.T.; Keeling, P.J. Lateral gene transfer and the complex distribution of insertions in eukaryotic enolase. Gene 2004, 340, 227–235. [Google Scholar] [CrossRef]
- Joint Genome Institute. Available online: www.jgi.doe.gov (accessed on 31 July 2014).
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Marechal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–251. [Google Scholar] [CrossRef]
- Prochnik, S.E.; Umen, J.; Nedelcu, A.M.; Hallmann, A.; Miller, S.M.; Nishii, I.; Ferris, P.; Kuo, A.; Mitros, T.; Fritz-Laylin, L.K.; et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 2010, 329, 223–226. [Google Scholar]
- Blanc, G.; Duncan, G.; Agarkova, I.; Borodovsky, M.; Gurnon, J.; Kuo, A.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Polle, J.; et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 2010, 22, 2943–2955. [Google Scholar] [CrossRef]
- Blanc, G.; Agarkova, I.; Grimwood, J.; Kuo, A.; Brueggeman, A.; Dunigan, D.D.; Gurnon, J.; Ladunga, I.; Lindquist, E.; Lucas, S.; et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012, 13, R39. [Google Scholar] [CrossRef]
- Worden, A.Z.; Lee, J.H.; Mock, T.; Rouze, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar]
- Palenik, B.; Grimwood, J.; Aerts, A.; Rouze, P.; Salamov, A.; Putnam, N.; Dupont, C.; Jorgensen, R.; Derelle, E.; Rombauts, S.; et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Nat. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar]
- Tardif, M.; Atteia, A.; Specht, M.; Cogne, G.; Rolland, N.; Brugiere, S.; Hippler, M.; Ferro, M.; Bruley, C.; Peltier, G.; et al. Predalgo: A new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012, 29, 3625–3639. [Google Scholar]
- Zhang Lab (I-TASSER ONLINE). Available online: http://zhanglab.ccmb.med.umich.edu/I-TASSER/ (accessed on 31 July 2014).
- Duan, J.; Li, S.; Ruan, K.; Bai, L. Cloning of promoter of the enolase gene from Dunaliella salina and its trasncription response to stress. Chin. J. Appl. Environ. Biol. 2011, 17, 202–206. [Google Scholar]
- Mettler, T.; Mühlhaus, T.; Hemme, D.; Schöttler, M.A.; Rupprecht, J.; Idoine, A.; Veyel, D.; Pal, S.K.; Yaneva-Roder, L.; Winck, F.V.; et al. Systems analysis of the response of photosynthesis, metabolism, and growth to an increase in irradiance in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell. 2014. [CrossRef]
- JGI Genome Portal. Available online: http://genome.jgi.doe.gov (accessed on 31 July 2014).
- Phytozome. Available online: www.phytozome.net (accessed on 31 July 2014).
- Geneious. Available online: www.geneious.com (accessed on 31 July 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Polle, J.E.W.; Neofotis, P.; Huang, A.; Chang, W.; Sury, K.; Wiech, E.M. Carbon Partitioning in Green Algae (Chlorophyta) and the Enolase Enzyme. Metabolites 2014, 4, 612-628. https://doi.org/10.3390/metabo4030612
Polle JEW, Neofotis P, Huang A, Chang W, Sury K, Wiech EM. Carbon Partitioning in Green Algae (Chlorophyta) and the Enolase Enzyme. Metabolites. 2014; 4(3):612-628. https://doi.org/10.3390/metabo4030612
Chicago/Turabian StylePolle, Jürgen E. W., Peter Neofotis, Andy Huang, William Chang, Kiran Sury, and Eliza M. Wiech. 2014. "Carbon Partitioning in Green Algae (Chlorophyta) and the Enolase Enzyme" Metabolites 4, no. 3: 612-628. https://doi.org/10.3390/metabo4030612