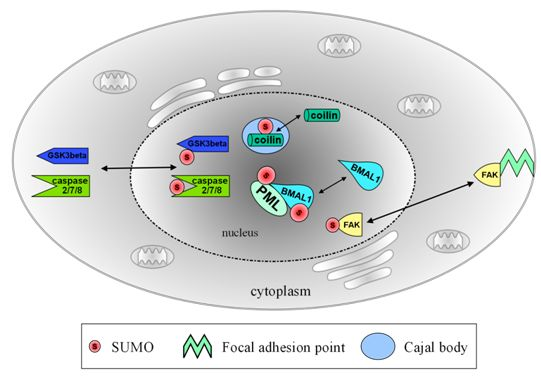
Regulation of Neuronal Protein Trafficking and Translocation by SUMOylation
Abstract
:1. Introduction
| Cellular compartment | SUMO substrate | Outcome of SUMOylation |
|---|---|---|
| intranuclear | PML | formation of PML bodies, possible transcriptional regulation upon axonal damage [9,10] |
| BMAL1 | association with PML bodies, transcriptional regulation and periodic degradation of BMAL1 [11,12] | |
| nucleo-cytoplasmic | GSK3beta | re-localization into the nucleus, enhances stability and stimulates apoptosis [13] |
| Caspase 2/7/8 | re-localization into the nucleus, possible cleavage of target proteins [14,15,16] | |
| FAK | re-localization into the nucleus, no functional data yet [17] | |
| extranuclear | Arc/Arg3.1 | Re-localization into dendrites and to cytoskeleton, important for establishment and maintenance of LTP [18] |
| GluK2 | internalization of receptor, possible recycling and re-insertion into plasma membrane [19] | |
| Group III mGluRs | Possible effects on synaptic transmission in the hippocampus, internalization and/or degradation of receptors [20,21] | |
| CB1 | Agonist-induced deSUMOylation potentially regulates internalization of receptor [22] | |
| La | binding to dynein and retrograde axonal transport [23] |
2. Regulation of Intra-Nuclear Organization by SUMO

2.1. Transient Localization of SUMO and Ubc9 at Cajal Bodies
2.2. PML Bodies and Axonal Damage
2.3. Keeping the Circadian Rhythm Running: SUMOylation of BMAL1
3. Extranuclear SUMOylation
3.1. AMPA Receptors, Arc/Arg3.1 and SUMOylation–a Possible Pathway for Induction of LTP and Synaptic Scaling
3.2. SUMOylation in Agonist-Induced Endocytosis and Plasticity of Kainate Receptors
3.3. Group III Metabotropic Glutamate Receptors–Genuine SUMO Targets?

3.4. G-Protein Coupled Cannabinoid Receptor 1
3.5. SUMOylation of La–Determining the Direction of Transport on the Microtubule Network
4. SUMOylation in Cytoplasm–Nuclear Transport
4.1. SUMOylation of Glycogen Synthase Kinase 3 β (GSK3β)–Translocation to the Nucleus
4.2. SUMO-Associated Nuclear Shuttling of Focal Adhesion Kinase (FAK)
4.3. Caspase SUMOylation Functions as a Nuclear Localization Signal
5. Conclusions
Acknowledgments
References
- Hannoun, Z.; Greenhough, S.; Jaffray, E.; Hay, R.T.; Hay, D.C. Post-translational modification by SUMO. Toxicology 2010, 278, 288–293. [Google Scholar]
- Praefcke, G.J.; Hofmann, K.; Dohmen, R.J. SUMO playing tag with ubiquitin. Trends Biochem. Sci. 2012, 37, 23–31. [Google Scholar]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar]
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 1997, 88, 97–107. [Google Scholar]
- Wilkinson, K.A.; Henley, J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010, 428, 133–145. [Google Scholar]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010, 11, 861–871. [Google Scholar]
- Craig, T.J.; Henley, J.M. Protein SUMOylation in spine structure and function. Curr. Opin. Neurobiol. 2011. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Nakamura, Y.; Henley, J.M. Targets and consequences of protein SUMOylation in neurons. Brain Res. Rev. 2010, 64, 195–212. [Google Scholar]
- Ishov, A.M.; Sotnikov, A.G.; Negorev, D.; Vladimirova, O.V.; Neff, N.; Kamitani, T.; Yeh, E.T.; Strauss, J.F., 3rd; Maul, G.G. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999, 147, 221–234. [Google Scholar] [CrossRef]
- Shen, T.H.; Lin, H.K.; Scaglioni, P.P.; Yung, T.M.; Pandolfi, P.P. The mechanisms of PML-nuclear body formation. Mol. Cell 2006, 24, 331–339. [Google Scholar]
- Lee, J.; Lee, Y.; Lee, M.J.; Park, E.; Kang, S.H.; Chung, C.H.; Lee, K.H.; Kim, K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell Biol. 2008, 28, 6056–6065. [Google Scholar]
- Cardone, L.; Hirayama, J.; Giordano, F.; Tamaru, T.; Palvimo, J.J.; Sassone-Corsi, P. Circadian clock control by SUMOylation of BMAL1. Science 2005, 309, 1390–1394. [Google Scholar]
- Eun Jeoung, L.; Sung Hee, H.; Jaesun, C.; Sung Hwa, S.; Kwang Hum, Y.; Min Kyoung, K.; Tae Yoon, P.; Sang Sun, K. Regulation of glycogen synthase kinase 3beta functions by modification of the small ubiquitin-like modifier. Open Biochem J. 2008, 2, 67–76. [Google Scholar]
- Besnault-Mascard, L.; Leprince, C.; Auffredou, M.T.; Meunier, B.; Bourgeade, M.F.; Camonis, J.; Lorenzo, H.K.; Vazquez, A. Caspase-8 sumoylation is associated with nuclear localization. Oncogene 2005, 24, 3268–3273. [Google Scholar]
- Hayashi, N.; Shirakura, H.; Uehara, T.; Nomura, Y. Relationship between SUMO-1 modification of caspase-7 and its nuclear localization in human neuronal cells. Neurosci. Lett. 2006, 397, 5–9. [Google Scholar]
- Shirakura, H.; Hayashi, N.; Ogino, S.; Tsuruma, K.; Uehara, T.; Nomura, Y. Caspase recruitment domain of procaspase-2 could be a target for SUMO-1 modification through Ubc9. Biochem. Biophys. Res. Commun. 2005, 331, 1007–1015. [Google Scholar]
- Kadare, G.; Toutant, M.; Formstecher, E.; Corvol, J.C.; Carnaud, M.; Boutterin, M.C.; Girault, J.A. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J. Biol. Chem. 2003, 278, 47434–47440. [Google Scholar]
- Bramham, C.R.; Alme, M.N.; Bittins, M.; Kuipers, S.D.; Nair, R.R.; Pai, B.; Panja, D.; Schubert, M.; Soule, J.; Tiron, A.; Wibrand, K. The Arc of synaptic memory. Exp. Brain Res. 2010, 200, 125–140. [Google Scholar]
- Martin, S.; Nishimune, A.; Mellor, J.R.; Henley, J.M. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature 2007, 447, 321–325. [Google Scholar]
- Dutting, E.; Schroder-Kress, N.; Sticht, H.; Enz, R. SUMO E3 ligases are expressed in the retina and regulate SUMOylation of the metabotropic glutamate receptor 8b. Biochem. J. 2011, 435, 365–371. [Google Scholar]
- Tang, Z.; El Far, O.; Betz, H.; Scheschonka, A. Pias1 interaction and sumoylation of metabotropic glutamate receptor 8. J. Biol. Chem. 2005, 280, 38153–38159. [Google Scholar]
- Gowran, A.; Murphy, C.E.; Campbell, V.A. Delta(9)-tetrahydrocannabinol regulates the p53 post-translational modifiers Murine double minute 2 and the Small Ubiquitin MOdifier protein in the rat brain. FEBS Lett. 2009, 583, 3412–3418. [Google Scholar]
- van Niekerk, E.A.; Willis, D.E.; Chang, J.H.; Reumann, K.; Heise, T.; Twiss, J.L. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc. Natl. Acad. Sci. USA 2007, 104, 12913–12918. [Google Scholar]
- Girdwood, D.W.; Tatham, M.H.; Hay, R.T. SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 2004, 15, 201–210. [Google Scholar]
- Dou, H.; Huang, C.; Van Nguyen, T.; Lu, L.S.; Yeh, E.T. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 2011, 585, 2891–2896. [Google Scholar]
- Watts, F.Z. Sumoylation of PCNA: Wrestling with recombination at stalled replication forks. DNA Repair (Amst) 2006, 5, 399–403. [Google Scholar] [CrossRef]
- Morris, G.E. The Cajal body. Biochim. Biophys. Acta 2008, 1783, 2108–2115. [Google Scholar]
- Berciano, M.T.; Novell, M.; Villagra, N.T.; Casafont, I.; Bengoechea, R.; Val-Bernal, J.F.; Lafarga, M. Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J. Struct. Biol. 2007, 158, 410–420. [Google Scholar]
- Navascues, J.; Bengoechea, R.; Tapia, O.; Casafont, I.; Berciano, M.T.; Lafarga, M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J. Struct. Biol. 2008, 163, 137–146. [Google Scholar]
- Sun, J.; Xu, H.; Subramony, S.H.; Hebert, M.D. Interactions between coilin and PIASy partially link Cajal bodies to PML bodies. J. Cell Sci. 2005, 118, 4995–5003. [Google Scholar]
- Watanabe, M.; Takahashi, K.; Tomizawa, K.; Mizusawa, H.; Takahashi, H. Developmental regulation of Ubc9 in the rat nervous system. Acta Biochim. Pol. 2008, 55, 681–686. [Google Scholar]
- Yan, Q.; Gong, L.; Deng, M.; Zhang, L.; Sun, S.; Liu, J.; Ma, H.; Yuan, D.; Chen, P.C.; Hu, X.; Qin, J.; Xiao, L.; Huang, X.Q.; Zhang, J.; Li, D.W. Sumoylation activates the transcriptional activity of Pax-6, an important transcription factor for eye and brain development. Proc. Natl. Acad. Sci. USA 2010, 107, 21034–21039. [Google Scholar]
- Shalizi, A.; Bilimoria, P.M.; Stegmuller, J.; Gaudilliere, B.; Yang, Y.; Shuai, K.; Bonni, A. PIASx is a MEF2 SUMO E3 ligase that promotes postsynaptic dendritic morphogenesis. J. Neurosci. 2007, 27, 10037–10046. [Google Scholar]
- Lin, D.Y.; Huang, Y.S.; Jeng, J.C.; Kuo, H.Y.; Chang, C.C.; Chao, T.T.; Ho, C.C.; Chen, Y.C.; Lin, T.P.; Fang, H.I.; Hung, C.C.; Suen, C.S.; Hwang, M.J.; Chang, K.S.; Maul, G.G.; Shih, H.M. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 2006, 24, 341–354. [Google Scholar]
- Tavalai, N.; Stamminger, T. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 2008, 1783, 2207–2221. [Google Scholar]
- Bernardi, R.; Papa, A.; Pandolfi, P.P. Regulation of apoptosis by PML and the PML-NBs. Oncogene 2008, 27, 6299–6312. [Google Scholar]
- Dellaire, G.; Bazett-Jones, D.P. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 2004, 26, 963–977. [Google Scholar]
- Lam, Y.W.; Ammerlaan, W.; O, W.S.; Kroese, F.; Opstelten, D. Cell type- and differentiation stage-dependent expression of PML domains in rat, detected by monoclonal antibody HIS55. Exp. Cell Res. 1995, 221, 344–356. [Google Scholar]
- Negorev, D.; Maul, G.G. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 2001, 20, 7234–7242. [Google Scholar]
- Villagra, N.T.; Berciano, J.; Altable, M.; Navascues, J.; Casafont, I.; Lafarga, M.; Berciano, M.T. PML bodies in reactive sensory ganglion neurons of the Guillain-Barre syndrome. Neurobiol. Dis. 2004, 16, 158–168. [Google Scholar]
- Griffin, J.W.; Li, C.Y.; Ho, T.W.; Tian, M.; Gao, C.Y.; Xue, P.; Mishu, B.; Cornblath, D.R.; Macko, C.; McKhann, G.M.; Asbury, A.K. Pathology of the motor-sensory axonal Guillain-Barre syndrome. Ann. Neurol. 1996, 39, 17–28. [Google Scholar]
- Tian, S.; Poukka, H.; Palvimo, J.J.; Janne, O.A. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem. J. 2002, 367, 907–911. [Google Scholar]
- Le Drean, Y.; Mincheneau, N.; Le Goff, P.; Michel, D. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology 2002, 143, 3482–3489. [Google Scholar]
- Chang, D.C.; Reppert, S.M. The circadian clocks of mice and men. Neuron 2001, 29, 555–558. [Google Scholar]
- Lamont, E.W.; Legault-Coutu, D.; Cermakian, N.; Boivin, D.B. The role of circadian clock genes in mental disorders. Dialogues Clin. Neurosci. 2007, 9, 333–342. [Google Scholar]
- Kiyohara, Y.B.; Tagao, S.; Tamanini, F.; Morita, A.; Sugisawa, Y.; Yasuda, M.; Yamanaka, I.; Ueda, H.R.; van der Horst, G.T.; Kondo, T.; Yagita, K. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci USA 2006, 103, 10074–10079. [Google Scholar]
- Perry, J.J.; Tainer, J.A.; Boddy, M.N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 2008, 33, 201–208. [Google Scholar]
- Martin, S.F.; Tatham, M.H.; Hay, R.T.; Samuel, I.D. Quantitative analysis of multi-protein interactions using FRET: application to the SUMO pathway. Protein Sci. 2008, 17, 777–784. [Google Scholar]
- Kessels, H.W.; Malinow, R. Synaptic AMPA receptor plasticity and behavior. Neuron 2009, 61, 340–350. [Google Scholar]
- Shepherd, J.D.; Huganir, R.L. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 2007, 23, 613–643. [Google Scholar]
- van der Sluijs, P.; Hoogenraad, C.C. New insights in endosomal dynamics and AMPA receptor trafficking. Semin. Cell Dev. Biol. 2011, 22, 499–505. [Google Scholar]
- Kantamneni, S.; Wilkinson, K.A.; Jaafari, N.; Ashikaga, E.; Rocca, D.; Rubin, P.; Jacobs, S.C.; Nishimune, A.; Henley, J.M. Activity-dependent SUMOylation of the brain-specific scaffolding protein GISP. Biochem. Biophys. Res. Commun. 2011, 409, 657–662. [Google Scholar]
- Chittajallu, R.; Braithwaite, S.P.; Clarke, V.R.; Henley, J.M. Kainate receptors: subunits, synaptic localization and function. Trends Pharmacol. Sci. 1999, 20, 26–35. [Google Scholar]
- Isaac, J.T.; Mellor, J.; Hurtado, D.; Roche, K.W. Kainate receptor trafficking: physiological roles and molecular mechanisms. Pharmacol. Ther. 2004, 104, 163–172. [Google Scholar]
- Lerma, J. Kainate receptor physiology. Curr. Opin. Pharmacol. 2006, 6, 89–97. [Google Scholar]
- Pinheiro, P.; Mulle, C. Kainate receptors. Cell Tissue Res. 2006, 326, 457–482. [Google Scholar]
- Martin, S.; Henley, J.M. Activity-dependent endocytic sorting of kainate receptors to recycling or degradation pathways. EMBO J. 2004, 23, 4749–4759. [Google Scholar]
- Konopacki, F.A.; Jaafari, N.; Rocca, D.L.; Wilkinson, K.A.; Chamberlain, S.; Rubin, P.; Kantamneni, S.; Mellor, J.R.; Henley, J.M. Agonist-induced PKC phosphorylation regulates GluK2 SUMOylation and kainate receptor endocytosis. Proc. Natl. Acad. Sci. USA 2011, 108, 19772–19777. [Google Scholar]
- Chamberlain, S.E.; Gonzalez-Gonzalez, I.M.; Wilkinson, K.A.; Konopacki, F.A.; Kantamneni, S.; Henley, J.M.; Mellor, J.R. SUMOylation and phosphorylation of GluK2 regulate kainate receptor trafficking and synaptic plasticity. Nat. Neurosci. 2012. [Google Scholar] [CrossRef]
- Shigemoto, R.; Kinoshita, A.; Wada, E.; Nomura, S.; Ohishi, H.; Takada, M.; Flor, P.J.; Neki, A.; Abe, T.; Nakanishi, S.; Mizuno, N. Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J. Neurosci. 1997, 17, 7503–7522. [Google Scholar]
- Wilkinson, K.A.; Henley, J.M. Analysis of metabotropic glutamate receptor 7 as a potential substrate for SUMOylation. Neurosci. Lett. 2011, 491, 181–186. [Google Scholar]
- Mackie, K. Distribution of cannabinoid receptors in the central and peripheral nervous system. Handb. Exp. Pharmacol. 2005, 299–325. [Google Scholar]
- Mackie, K. Cannabinoid receptors: where they are and what they do. J. Neuroendocrinol. 2008, 20 (Suppl. 1), 10–14. [Google Scholar] [CrossRef]
- Carter, S.; Bischof, O.; Dejean, A.; Vousden, K.H. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 2007, 9, 428–435. [Google Scholar]
- Twiss, J.L.; van Minnen, J. New insights into neuronal regeneration: the role of axonal protein synthesis in pathfinding and axonal extension. J. Neurotrauma. 2006, 23, 295–308. [Google Scholar]
- Hirokawa, N. mRNA transport in dendrites: RNA granules, motors, and tracks. J. Neurosci. 2006, 26, 7139–7142. [Google Scholar]
- Cardinali, B.; Carissimi, C.; Gravina, P.; Pierandrei-Amaldi, P. La protein is associated with terminal oligopyrimidine mRNAs in actively translating polysomes. J. Biol. Chem. 2003, 278, 35145–35151. [Google Scholar]
- Willis, D.; Li, K.W.; Zheng, J.Q.; Chang, J.H.; Smit, A.; Kelly, T.; Merianda, T.T.; Sylvester, J.; van Minnen, J.; Twiss, J.L. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci. 2005, 25, 778–791. [Google Scholar]
- Mishra, R. Glycogen synthase kinase 3 beta: can it be a target for oral cancer. Mol. Cancer 2010, 9, 144. [Google Scholar]
- Doble, B.W.; Woodgett, J.R. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003, 116, 1175–1186. [Google Scholar]
- Rayasam, G.V.; Tulasi, V.K.; Sodhi, R.; Davis, J.A.; Ray, A. Glycogen synthase kinase 3: more than a namesake. Br. J. Pharmacol. 2009, 156, 885–898. [Google Scholar]
- Hur, E.M.; Zhou, F.Q. GSK3 signalling in neural development. Nat. Rev. Neurosci. 2010, 11, 539–551. [Google Scholar]
- Peineau, S.; Taghibiglou, C.; Bradley, C.; Wong, T.P.; Liu, L.; Lu, J.; Lo, E.; Wu, D.; Saule, E.; Bouschet, T.; Matthews, P.; Isaac, J.T.; Bortolotto, Z.A.; Wang, Y.T.; Collingridge, G.L. LTP inhibits LTD in the hippocampus via regulation of GSK3beta. Neuron 2007, 53, 703–717. [Google Scholar]
- Mitra, S.K.; Hanson, D.A.; Schlaepfer, D.D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 2005, 6, 56–68. [Google Scholar]
- Hall, J.E.; Fu, W.; Schaller, M.D. Focal adhesion kinase: exploring Fak structure to gain insight into function. Int. Rev. Cell Mol. Biol. 2011, 288, 185–225. [Google Scholar]
- Valiente, M.; Ciceri, G.; Rico, B.; Marin, O. Focal adhesion kinase modulates radial glia-dependent neuronal migration through connexin-26. J. Neurosci. 2011, 31, 11678–11691. [Google Scholar]
- Nikolic, M. The molecular mystery of neuronal migration: FAK and Cdk5. Trends Cell Biol. 2004, 14, 1–5. [Google Scholar]
- Robles, E.; Gomez, T.M. Focal adhesion kinase signaling at sites of integrin-mediated adhesion controls axon pathfinding. Nat. Neurosci. 2006, 9, 1274–1283. [Google Scholar]
- Cobb, B.S.; Schaller, M.D.; Leu, T.H.; Parsons, J.T. Stable association of pp60src and pp59fyn with the focal adhesion-associated protein tyrosine kinase, pp125FAK. Mol. Cell Biol. 1994, 14, 147–155. [Google Scholar]
- Kotaja, N.; Karvonen, U.; Janne, O.A.; Palvimo, J.J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell Biol. 2002, 22, 5222–5234. [Google Scholar]
- Ribe, E.M.; Serrano-Saiz, E.; Akpan, N.; Troy, C.M. Mechanisms of neuronal death in disease: defining the models and the players. Biochem. J. 2008, 415, 165–182. [Google Scholar]
- Li, Z.; Jo, J.; Jia, J.M.; Lo, S.C.; Whitcomb, D.J.; Jiao, S.; Cho, K.; Sheng, M. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell 2010, 141, 859–871. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Berndt, A.; Wilkinson, K.A.; Henley, J.M. Regulation of Neuronal Protein Trafficking and Translocation by SUMOylation. Biomolecules 2012, 2, 256-268. https://doi.org/10.3390/biom2020256
Berndt A, Wilkinson KA, Henley JM. Regulation of Neuronal Protein Trafficking and Translocation by SUMOylation. Biomolecules. 2012; 2(2):256-268. https://doi.org/10.3390/biom2020256
Chicago/Turabian StyleBerndt, Anja, Kevin A. Wilkinson, and Jeremy M. Henley. 2012. "Regulation of Neuronal Protein Trafficking and Translocation by SUMOylation" Biomolecules 2, no. 2: 256-268. https://doi.org/10.3390/biom2020256