Functional Aspects of PARP1 in DNA Repair and Transcription
Abstract
:1. Introduction
2. PARP1 Function and Regulation
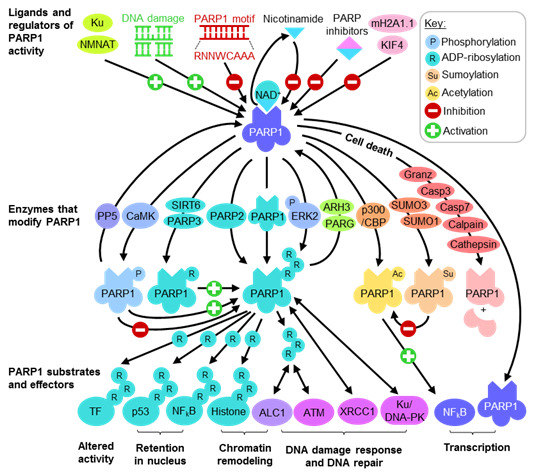
2.1. Regulating PARP1 ADP-Ribosylation Activity
2.2. PARP1 Substrates and Effectors Play Important Roles in Transcription and DNA Damage Response
3. PARP1 ADP-Ribosylation Activity Is Important for Mediating DNA Repair
| DNA Repair Mechanism | PARP1 Function | References |
|---|---|---|
| Base excision repair (BER) | Binds AP site | [87] |
| Auto-modified PARP1 recruits BER complex | [88] | |
| Nucleotide excision repair (NER) | ADP-ribosylates XPA | [89,90] |
| Mismatch repair (MMR) | ADP-ribosylates MSH6 | [89,90] |
| Single-strand break repair (SSBR) | Auto-modified PARP1 recruits BER complex | [89,91,92] |
| Double-strand break repair by nonhomologous end joining (NHEJ) | Ku enhances PARP1 ADP-ribosylation activity | [89,90] |
| ADP-ribosylates and activates DNA-PKcs | ||
| Double-strand break repair by homologous recombination (HR) | Auto-modified PARP1 recruits Mre11 | [66] |
| PAR activates ATM signalling | [74] |
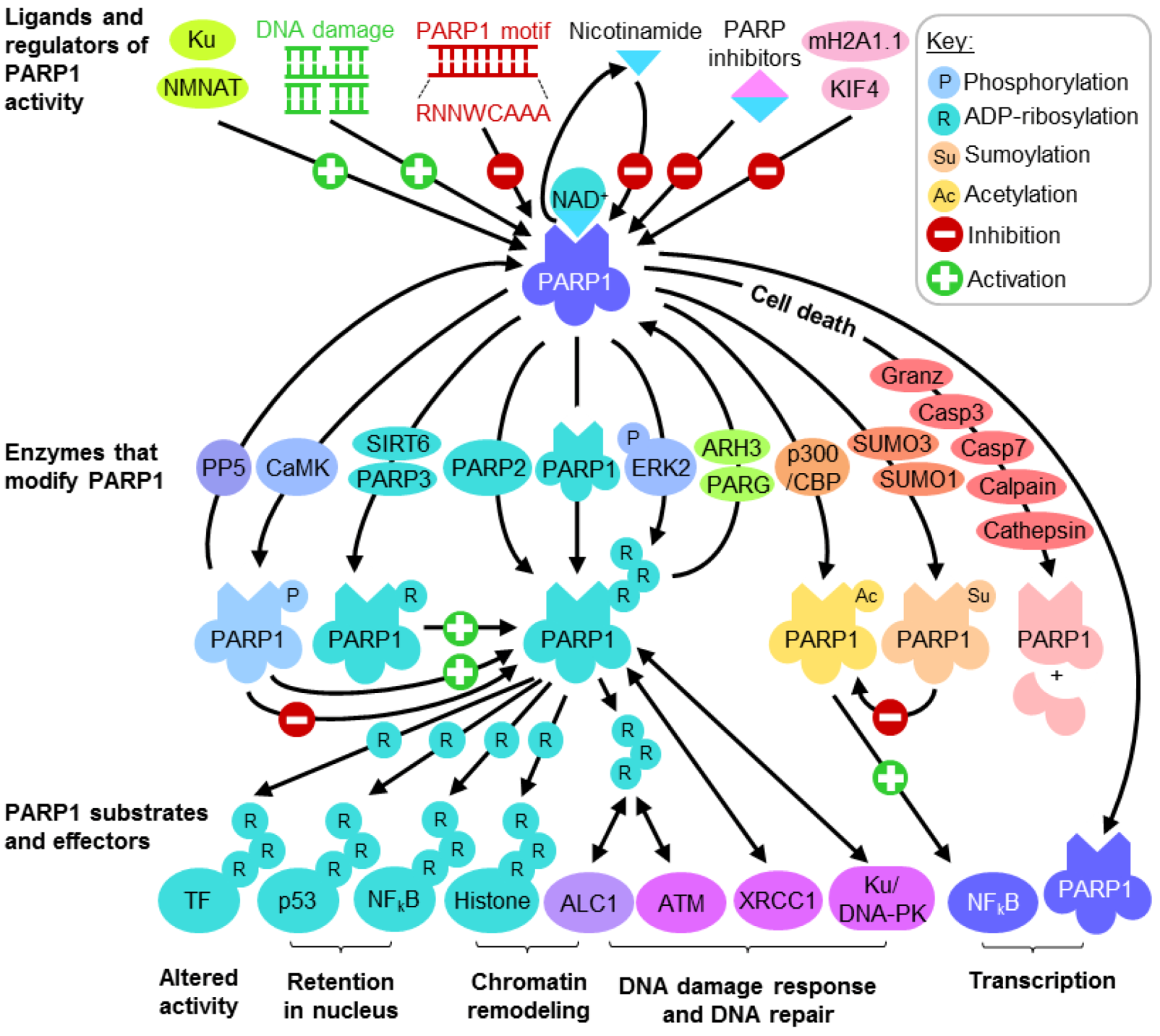
3.1. PARP1 in the Repair of Modified DNA
3.2. PARP1 in the Repair of DNA Strand Breaks
3.3. Uncontrolled PARP1 ADP-Ribosylation Activity during DNA Repair Results in Cell Death
4. PARP1 as a Transcriptional Regulator Controlling Expression of DNA Damage Response Genes
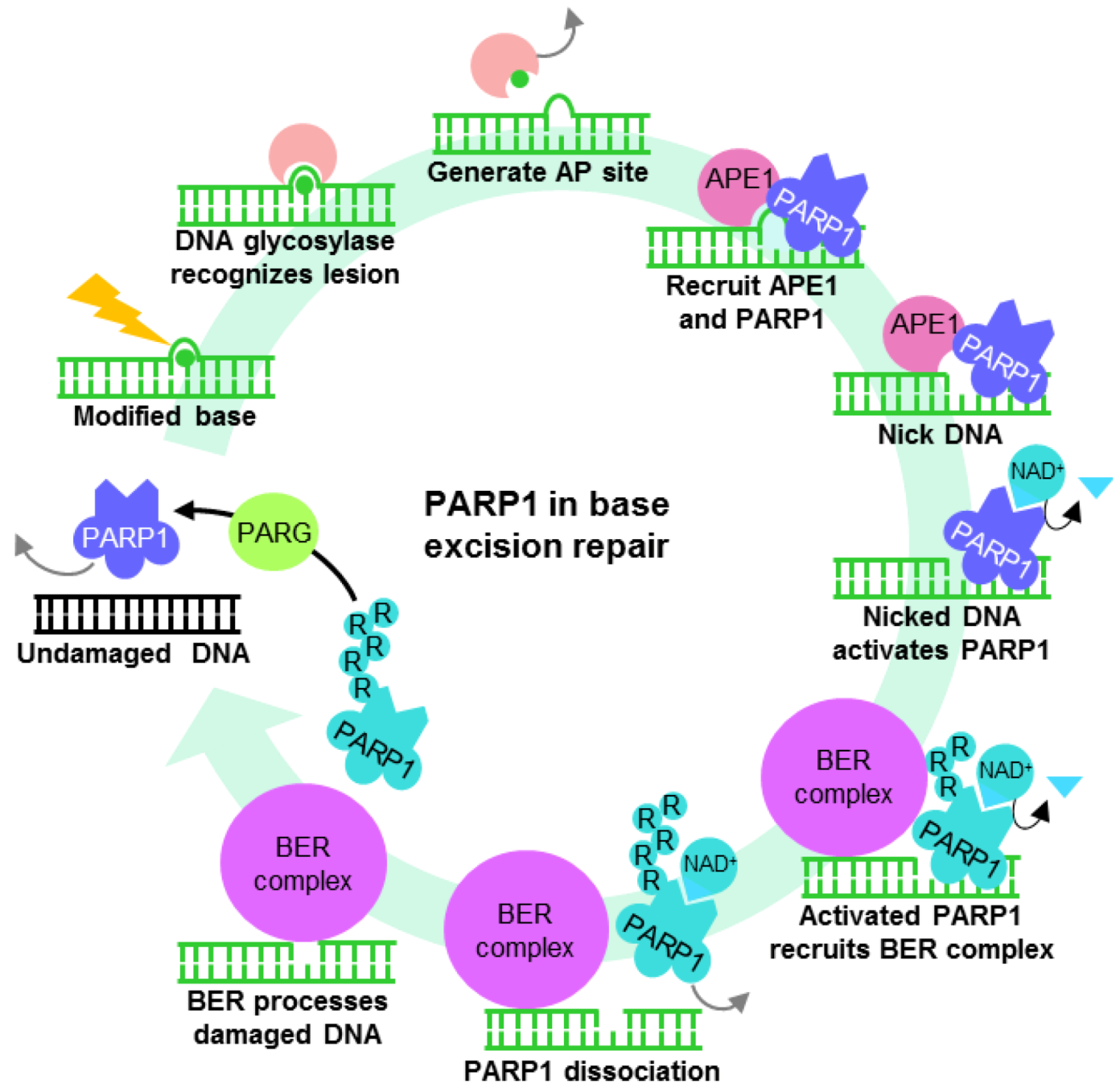
4.1. PARP1 ADP-Ribosylation Activity Controls Transcription States
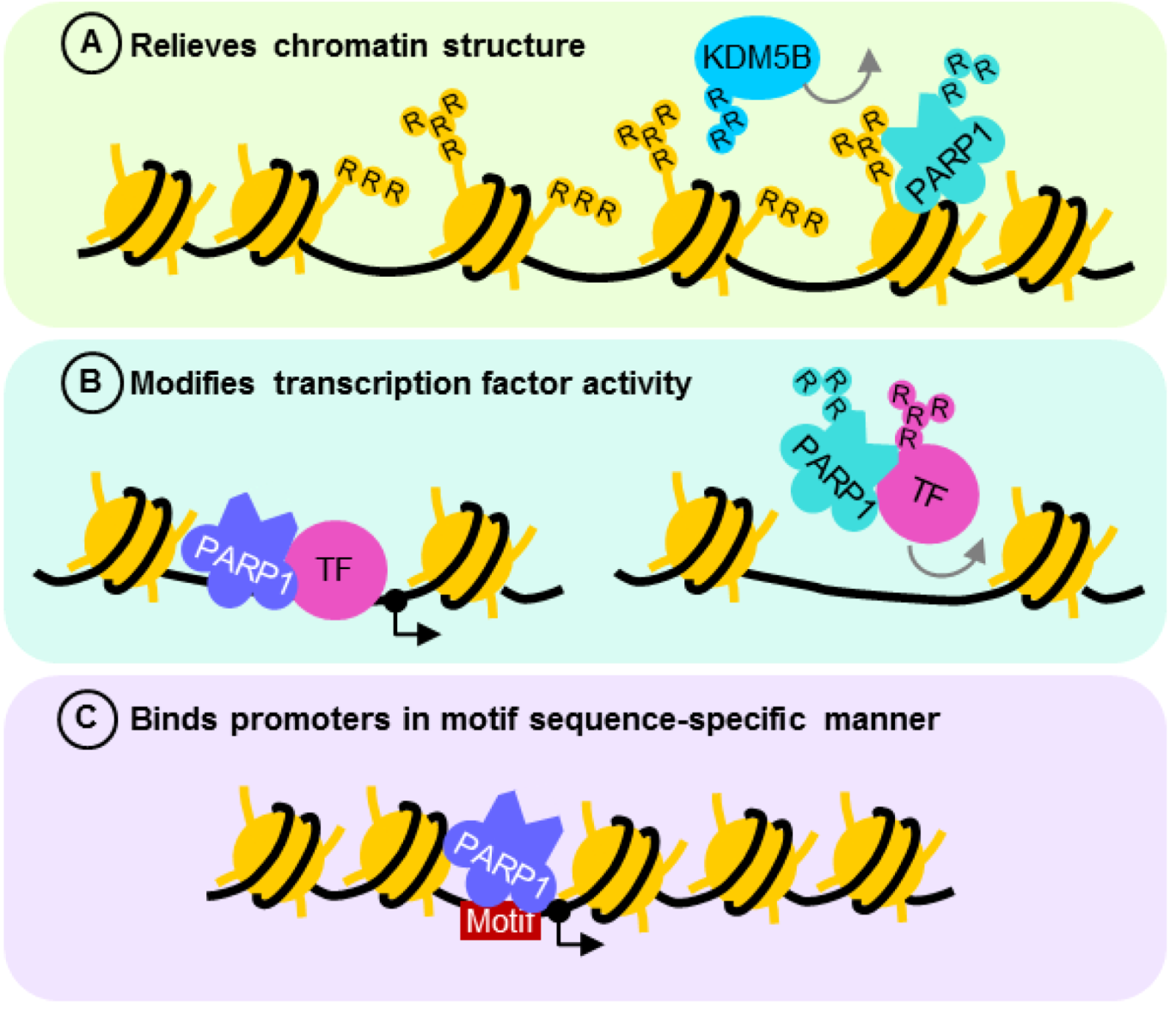
4.2. PARP1 is a Motif-Dependent Transcription Factor
| DNA repair mechanism | Gene | Gene function [References] | PARP1 motif |
|---|---|---|---|
| Double-strand break repair by homologous recombination (HR) | BRCA1 | E3 ubiquitin ligase with multiple roles including controlling DNA damage signaling [131] | GAAACAAA |
| BRCA2# | Mediates recombination [132,133] | GGTACAAA | |
| BRIP1 | Interacts with BRCA1 [131] | AGTTCAAA | |
| OBFC2B | SOSS complex component; ATM signaling [134] | GCGACAAA | |
| SSBIP1 | SOSS complex component; ATM signaling [134] | GAGACAAA | |
| TOPBP1 | Stalled replication forks; ATR signaling [135] | ATTTCAAA | |
| NSMCE2 | E3 SUMO ligase of SMC5-SMC6 complex [136] | GGATCAAA | |
| SLX1B | SLX1-SLX4 resolvase catalytic subunit [137,138] | AGGACAAA | |
| DMC1 | Meiosis-specific recombinase; Interacts with BRCA2 [132,139] | AGAACAAA | |
| Base excision repair (BER) | NEIL3 | DNA glycosylase [140] | AGCTCAAA |
| MBD4^ | DNA glycosylase specific for G:T or G:U mismatches within CpG islands [141,142] | ACAACAAA | |
| Nucleotide excision repair (NER) | CETN2 | Component of XPC complex [143] | GAGACAAA |
| Mismatch repair (MMR) | MSH6 | Component of MMR [91] | GGGTCAAA |
| Direct base reversal | ALKBH3 | Oxidative demethylation of alkylated DNA [144,145] | GCCACAAA |
| Interstrand crosslink repair (ICL) | FANCG | Component of FA core complex [146] | ACTACAAA |
| DNA repair accessory proteins | RPA1 | Stabilize single-strand DNA intermediates | GTGACAAA |
| DNA polymerases | POLA2 | Subunit of primase complex | GCTACAAA |
| POLD3 | DNA polymerase δ subunit | ACTTCAAA |
5. Dysregulated PARP1 ADP-Ribosylation and Transcription Activities

5.1. PARP1 Inhibition, Enzymatic Hyper-Activation and Disease
5.2. Oncogenic Viruses—Inhibiting the PARP1 Enzyme to Enhance Viral Replication
| Oncogenic virus | Gene or DNA element | Motif |
|---|---|---|
| Human herpesvirus 4 (EBV) | OriLyt replication origin | ACTTCAAA |
| Hepatitis B Virus (HBV) | Core promoter | ACTTCAAA |
| Human T-cell leukemia virus (HTLV) | Tax responsive element | ACGACAAC |
| Human herpesvirus 8 (KSHV) | ORF4 complement control protein | GCTACAAA |
| Primase | ACGTCAAA | |
| Merkel cell polyomavirus | VP3 capsid protein | ACTTCAAA |
6. Conclusions
Conflict of Interest
Acknowledgments
References
- Satoh, M.S.; Poirier, G.G.; Lindahl, T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry 1994, 33, 7099–7106. [Google Scholar]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011, 286, 10690–10701. [Google Scholar]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Eustermann, S.; Videler, H.; Yang, J.C.; Cole, P.T.; Gruszka, D.; Veprintsev, D.; Neuhaus, D. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 2011, 407, 149–170. [Google Scholar] [CrossRef]
- Lindahl, T.; Satoh, M.S.; Poirier, G.G.; Klungland, A. Post-translational modification of poly(adp-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995, 20, 405–411. [Google Scholar] [CrossRef]
- D'Amours, D.; Desnoyers, S.; D'Silva, I.; Poirier, G.G. Poly(adp-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342 (Pt 2), 249–268. [Google Scholar]
- De Vos, M.; Schreiber, V.; Dantzer, F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem. Pharmacol. 2012, 84, 137–146. [Google Scholar]
- Jagtap, P.; Szabo, C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat. Rev. Drug Discov. 2005, 4, 421–440. [Google Scholar] [CrossRef]
- Rouleau, M.; Patel, A.; Hendzel, M.J.; Kaufmann, S.H.; Poirier, G.G. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer 2010, 10, 293–301. [Google Scholar] [CrossRef]
- Shall, S.; de Murcia, G. Poly(ADP-ribose) polymerase-1: What have we learned from the deficient mouse model? Mutat. Res. 2000, 460, 1–15. [Google Scholar] [CrossRef]
- Masutani, M.; Nakagama, H.; Sugimura, T. Poly(ADP-ribosyl)ation in relation to cancer and autoimmune disease. Cell. Mol. Life Sci. 2005, 62, 769–783. [Google Scholar] [CrossRef]
- Szabo, C.; Pacher, P.; Swanson, R.A. Novel modulators of poly(ADP-ribose) polymerase. Trends Pharmacol. Sci. 2006, 27, 626–630. [Google Scholar] [CrossRef]
- Miwa, M.; Masutani, M. PolyADP-ribosylation and cancer. Cancer Sci. 2007, 98, 1528–1535. [Google Scholar] [CrossRef]
- Kling, J. PARP inhibitors blaze a trail in difficult-to-treat cancers. Nat. Biotechnol. 2009, 27, 784–786. [Google Scholar] [CrossRef]
- Peralta-Leal, A.; Rodriguez-Vargas, J.M.; Aguilar-Quesada, R.; Rodriguez, M.I.; Linares, J.L.; de Almodovar, M.R.; Oliver, F.J. PARP inhibitors: New partners in the therapy of cancer and inflammatory diseases. Free Radic. Biol. Med. 2009, 47, 13–26. [Google Scholar] [CrossRef]
- Schreiber, V.; Dantzer, F.; Ame, J.C.; de Murcia, G. Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006, 7, 517–528. [Google Scholar] [CrossRef]
- Plummer, E.R. Inhibition of poly(ADP-ribose) polymerase in cancer. Curr. Opin. Pharmacol. 2006, 6, 364–368. [Google Scholar] [CrossRef]
- Kim, M.Y.; Mauro, S.; Gevry, N.; Lis, J.T.; Kraus, W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 2004, 119, 803–814. [Google Scholar] [CrossRef]
- Kraus, W.L.; Lis, J.T. PARP goes transcription. Cell 2003, 113, 677–683. [Google Scholar] [CrossRef]
- Kraus, W.L. Transcriptional control by PARP-1: Chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008, 20, 294–302. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kraus, W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef]
- Rouleau, M.; Aubin, R.A.; Poirier, G.G. Poly(ADP-ribosyl)ated chromatin domains: Access granted. J. Cell Sci. 2004, 117, 815–825. [Google Scholar] [CrossRef]
- Ko, H.L.; Ren, E.C. Novel poly (ADP-ribose) polymerase 1 binding motif in hepatitis B virus core promoter impairs DNA damage repair. Hepatology 2011, 54, 1190–1198. [Google Scholar] [CrossRef]
- Zhang, Z.; Hildebrandt, E.F.; Simbulan-Rosenthal, C.M.; Anderson, M.G. Sequence-specific binding of poly(ADP-ribose) polymerase-1 to the human T cell leukemia virus type-I tax responsive element. Virology 2002, 296, 107–116. [Google Scholar] [CrossRef]
- Ambrose, H.E.; Papadopoulou, V.; Beswick, R.W.; Wagner, S.D. Poly-(ADP-ribose) polymerase-1 (PARP-1) binds in a sequence-specific manner at the bcl-6 locus and contributes to the regulation of bcl-6 transcription. Oncogene 2007, 26, 6244–6252. [Google Scholar] [CrossRef]
- Pottier, N.; Cheok, M.H.; Yang, W.; Assem, M.; Tracey, L.; Obenauer, J.C.; Panetta, J.C.; Relling, M.V.; Evans, W.E. Expression of SMARCB1 modulates steroid sensitivity in human lymphoblastoid cells: Identification of a promoter SNP that alters PARP1 binding and SMARCB1 expression. Hum. Mol. Genet. 2007, 16, 2261–2271. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, J.D.; Poon, V.K.; Chen, D.Q.; Chan, C.C.; Ng, F.; Guan, X.Y.; Watt, R.M.; Lu, L.; Yuen, K.Y., et al. Functional dissection of an IFN-alpha/beta receptor 1 promoter variant that confers higher risk to chronic hepatitis b virus infection. J. Hepatol. 2009, 51, 322–332. [Google Scholar]
- Chung, E.Y.; Liu, J.; Zhang, Y.; Ma, X. Differential expression in lupus-associated Il-10 promoter single-nucleotide polymorphisms is mediated by poly(ADP-ribose) polymerase-1. Genes Immun. 2007, 8, 577–589. [Google Scholar] [CrossRef]
- Akiyama, T.; Takasawa, S.; Nata, K.; Kobayashi, S.; Abe, M.; Shervani, N.J.; Ikeda, T.; Nakagawa, K.; Unno, M.; Matsuno, S., et al. Activation of Reg gene, a gene for insulin-producing beta-cell regeneration: Poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by auto poly(ADP-ribosyl)ation. Proc. Natl. Acad. Sci. USA 2001, 98, 48–53. [Google Scholar]
- Nyquist, P.; Zhang, J.; De Graba, T.J. The -928 G/C and -362 G/C single-nucleotide polymorphisms in the promoter of MCP-1: Increased transcriptional activity and novel binding sites. Cerebrovasc. Dis. 2009, 29, 242–247. [Google Scholar]
- Hassa, P.O.; Haenni, S.S.; Elser, M.; Hottiger, M.O. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006, 70, 789–829. [Google Scholar] [CrossRef]
- Pion, E.; Ullmann, G.M.; Ame, J.C.; Gerard, D.; de Murcia, G.; Bombarda, E. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry 2005, 44, 14670–14681. [Google Scholar]
- Petrucco, S.; Percudani, R. Structural recognition of DNA by poly(ADP-ribose)polymerase-like zinc finger families. FEBS J. 2008, 275, 883–893. [Google Scholar] [CrossRef]
- Langelier, M.F.; Ruhl, D.D.; Planck, J.L.; Kraus, W.L.; Pascal, J.M. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010, 285, 18877–18887. [Google Scholar]
- Gagne, J.P.; Rouleau, M.; Poirier, G.G. Structural biology. PARP-1 activation--bringing the pieces together. Science 2010, 336, 678–679. [Google Scholar]
- Luo, X.; Kraus, W.L. On par with parp: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Hong, S.J.; Dawson, T.M.; Dawson, V.L. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol. Sci. 2004, 25, 259–264. [Google Scholar] [CrossRef]
- Huber, A.; Bai, P.; de Murcia, J.M.; de Murcia, G. PARP-1, PARP-2 and ATM in the DNA damage response: Functional synergy in mouse development. DNA Repair (Amst) 2004, 3, 1103–1108. [Google Scholar]
- Kim, M.Y.; Zhang, T.; Kraus, W.L. Poly(ADP-ribosyl)ation by PARP-1: 'Par-laying' NAD+ into a nuclear signal. Genes Dev. 2005, 19, 1951–1967. [Google Scholar] [CrossRef]
- Koh, D.W.; Dawson, T.M.; Dawson, V.L. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol. Res. 2005, 52, 5–14. [Google Scholar] [CrossRef]
- Heeres, J.T.; Hergenrother, P.J. Poly(ADP-ribose) makes a date with death. Curr. Opin. Chem. Biol. 2007, 11, 644–653. [Google Scholar] [CrossRef]
- Hakme, A.; Wong, H.K.; Dantzer, F.; Schreiber, V. The expanding field of poly(ADP-ribosyl)ation reactions. 'Protein modifications: Beyond the usual suspects' review series. EMBO Rep. 2008, 9, 1094–1100. [Google Scholar]
- Ogata, N.; Ueda, K.; Kawaichi, M.; Hayaishi, O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 1981, 256, 4135–4137. [Google Scholar]
- Ame, J.C.; Rolli, V.; Schreiber, V.; Niedergang, C.; Apiou, F.; Decker, P.; Muller, S.; Hoger, T.; Menissier-de Murcia, J.; de Murcia, G. Parp-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999, 274, 17860–17868. [Google Scholar]
- Koh, D.W.; Lawler, A.M.; Poitras, M.F.; Sasaki, M.; Wattler, S.; Nehls, M.C.; Stoger, T.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 2004, 101, 17699–17704. [Google Scholar]
- Loseva, O.; Jemth, A.S.; Bryant, H.E.; Schuler, H.; Lehtio, L.; Karlberg, T.; Helleday, T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J. Biol. Chem. 2010, 285, 8054–8060. [Google Scholar]
- Mao, Z.; Hine, C.; Tian, X.; Meter, M. V.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef]
- Kauppinen, T.M.; Chan, W.Y.; Suh, S.W.; Wiggins, A.K.; Huang, E.J.; Swanson, R.A. Direct phosphorylation and regulation of poly(ADP-ribose) polymerase-1 by extracellular signal-regulated kinases 1/2. Proc. Natl. Acad. Sci. USA 2006, 103, 7136–7141. [Google Scholar]
- Cohen-Armon, M.; Visochek, L.; Rozensal, D.; Kalal, A.; Geistrikh, I.; Klein, R.; Bendetz-Nezer, S.; Yao, Z.; Seger, R. DNA-independent PARP-1 activation by phosphorylated ERK2 increases ELK1 activity: A link to histone acetylation. Mol. Cell 2007, 25, 297–308. [Google Scholar] [CrossRef]
- Cohen-Armon, M. PARP-1 activation in the ERK signaling pathway. Trends Pharmacol. Sci. 2007, 28, 556–560. [Google Scholar] [CrossRef]
- Ju, B.G.; Solum, D.; Song, E.J.; Lee, K.J.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a camkinase II delta-dependent neurogenic gene activation pathway. Cell 2004, 119, 815–829. [Google Scholar] [CrossRef]
- Dong, F.; Soubeyrand, S.; Hache, R.J. Activation of PARP-1 in response to bleomycin depends on the Ku antigen and protein phosphatase 5. Oncogene 2010, 29, 2093–2103. [Google Scholar] [CrossRef]
- Messner, S.; Schuermann, D.; Altmeyer, M.; Kassner, I.; Schmidt, D.; Schar, P.; Muller, S.; Hottiger, M.O. Sumoylation of poly(ADP-ribose) polymerase 1 inhibits its acetylation and restrains transcriptional coactivator function. FASEB J. 2009, 23, 3978–3989. [Google Scholar] [CrossRef] [Green Version]
- Berger, F.; Lau, C.; Ziegler, M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear nad biosynthetic enzyme NMN adenylyl transferase 1. Proc. Natl. Acad. Sci. USA 2007, 104, 3765–3770. [Google Scholar]
- Ouararhni, K.; Hadj-Slimane, R.; Ait-Si-Ali, S.; Robin, P.; Mietton, F.; Harel-Bellan, A.; Dimitrov, S.; Hamiche, A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006, 20, 3324–3336. [Google Scholar]
- Midorikawa, R.; Takei, Y.; Hirokawa, N. KIF4 motor regulates activity-dependent neuronal survival by suppressing PARP-1 enzymatic activity. Cell 2006, 125, 371–383. [Google Scholar] [CrossRef]
- Kaufmann, S.H.; Desnoyers, S.; Ottaviano, Y.; Davidson, N.E.; Poirier, G.G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: An early marker of chemotherapy-induced apoptosis. Cancer Res. 1993, 53, 3976–3985. [Google Scholar]
- Nicholson, D.W.; Ali, A.; Thornberry, N.A.; Vaillancourt, J.P.; Ding, C.K.; Gallant, M.; Gareau, Y.; Griffin, P.R.; Labelle, M.; Lazebnik, Y.A., et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 1995, 376, 37–43. [Google Scholar]
- Erdelyi, K.; Bakondi, E.; Gergely, P.; Szabo, C.; Virag, L. Pathophysiologic role of oxidative stress-induced poly(ADP-ribose) polymerase-1 activation: Focus on cell death and transcriptional regulation. Cell. Mol. Life Sci. 2005, 62, 751–759. [Google Scholar] [CrossRef]
- Chevanne, M.; Calia, C.; Zampieri, M.; Cecchinelli, B.; Caldini, R.; Monti, D.; Bucci, L.; Franceschi, C.; Caiafa, P. Oxidative DNA damage repair and PARP 1 and PARP 2 expression in epstein-barr virus-immortalized b lymphocyte cells from young subjects, old subjects, and centenarians. Rejuvenation Res. 2007, 10, 191–204. [Google Scholar] [CrossRef]
- Chaitanya, G.V.; Babu, P.P. Differential parp cleavage: An indication of heterogeneous forms of cell death and involvement of multiple proteases in the infarct of focal cerebral ischemia in rat. Cell. Mol. Neurobiol. 2009, 29, 563–573. [Google Scholar] [CrossRef]
- McGinnis, K.M.; Gnegy, M.E.; Park, Y.H.; Mukerjee, N.; Wang, K.K. Procaspase-3 and poly(ADP)ribose polymerase (PARP) are calpain substrates. Biochem. Biophys. Res. Commun. 1999, 263, 94–99. [Google Scholar] [CrossRef]
- Gobeil, S.; Boucher, C.C.; Nadeau, D.; Poirier, G.G. Characterization of the necrotic cleavage of poly(ADP-ribose) polymerase (PARP-1): Implication of lysosomal proteases. Cell Death Differ. 2001, 8, 588–594. [Google Scholar] [CrossRef]
- Hassa, P.O.; Haenni, S.S.; Buerki, C.; Meier, N.I.; Lane, W.S.; Owen, H.; Gersbach, M.; Imhof, R.; Hottiger, M.O. Acetylation of poly(ADP-ribose) polymerase-1 by p300/creb-binding protein regulates coactivation of NF-kappaB-dependent transcription. J. Biol. Chem. 2005, 280, 40450–40464. [Google Scholar]
- Ju, B.G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase II beta-mediated dsdna break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef]
- Bryant, H.E.; Petermann, E.; Schultz, N.; Jemth, A.S.; Loseva, O.; Issaeva, N.; Johansson, F.; Fernandez, S.; McGlynn, P.; Helleday, T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009, 28, 2601–2615. [Google Scholar] [CrossRef]
- Gottschalk, A.J.; Timinszky, G.; Kong, S.E.; Jin, J.; Cai, Y.; Swanson, S.K.; Washburn, M.P.; Florens, L.; Ladurner, A.G.; Conaway, J.W., et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. USA 2009, 106, 13770–13774. [Google Scholar]
- Stilmann, M.; Hinz, M.; Arslan, S.C.; Zimmer, A.; Schreiber, V.; Scheidereit, C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced Ikappab kinase activation. Mol. Cell 2009, 36, 365–378. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kraus, W.L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 2010, 39, 736–749. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Gamble, M.J.; Frizzell, K.M.; Berrocal, J.G.; Kininis, M.; Kraus, W.L. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science 2008, 319, 819–821. [Google Scholar]
- Timinszky, G.; Till, S.; Hassa, P.O.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.; Scheffzek, K., et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar]
- Masson, M.; Niedergang, C.; Schreiber, V.; Muller, S.; Menissier-de Murcia, J.; de Murcia, G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998, 18, 3563–3571. [Google Scholar]
- Ahel, D.; Horejsi, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P., et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef]
- Haince, J.F.; Kozlov, S.; Dawson, V.L.; Dawson, T.M.; Hendzel, M.J.; Lavin, M.F.; Poirier, G.G. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 2007, 282, 16441–16453. [Google Scholar]
- Smith, S. The world according to PARP. Trends Biochem. Sci. 2001, 26, 174–179. [Google Scholar] [CrossRef]
- Nie, J.; Sakamoto, S.; Song, D.; Qu, Z.; Ota, K.; Taniguchi, T. Interaction of oct-1 and automodification domain of poly(ADP-ribose) synthetase. FEBS Lett. 1998, 424, 27–32. [Google Scholar] [CrossRef]
- Gagne, J.P.; Hunter, J.M.; Labrecque, B.; Chabot, B.; Poirier, G.G. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 2003, 371, 331–340. [Google Scholar] [CrossRef]
- Zerfaoui, M.; Errami, Y.; Naura, A.S.; Suzuki, Y.; Kim, H.; Ju, J.; Liu, T.; Hans, C.P.; Kim, J.G.; Abd Elmageed, Z.Y., et al. Poly(ADP-ribose) polymerase-1 is a determining factor in crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J. Immunol. 2010, 185, 1894–1902. [Google Scholar]
- Kanai, M.; Hanashiro, K.; Kim, S.H.; Hanai, S.; Boulares, A.H.; Miwa, M.; Fukasawa, K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell. Biol. 2007, 9, 1175–1183. [Google Scholar] [CrossRef]
- Shibata, A.; Kamada, N.; Masumura, K.; Nohmi, T.; Kobayashi, S.; Teraoka, H.; Nakagama, H.; Sugimura, T.; Suzuki, H.; Masutani, M. PARP-1 deficiency causes an increase of deletion mutations and insertions/rearrangements in vivo after treatment with an alkylating agent. Oncogene 2005, 24, 1328–1337. [Google Scholar] [CrossRef]
- Shibata, A.; Maeda, D.; Ogino, H.; Tsutsumi, M.; Nohmi, T.; Nakagama, H.; Sugimura, T.; Teraoka, H.; Masutani, M. Role of PARP-1 in suppressing spontaneous deletion mutation in the liver and brain of mice at adolescence and advanced age. Mutat. Res. 2009, 664, 20–27. [Google Scholar] [CrossRef]
- Tong, W.M.; Cortes, U.; Hande, M.P.; Ohgaki, H.; Cavalli, L.R.; Lansdorp, P.M.; Haddad, B.R.; Wang, Z.Q. Synergistic role of Ku80 and poly(ADP-ribose) polymerase in suppressing chromosomal aberrations and liver cancer formation. Cancer Res. 2002, 62, 6990–6996. [Google Scholar]
- Tong, W.M.; Yang, Y.G.; Cao, W.H.; Galendo, D.; Frappart, L.; Shen, Y.; Wang, Z.Q. Poly(ADP-ribose) polymerase-1 plays a role in suppressing mammary tumourigenesis in mice. Oncogene 2007, 26, 3857–3867. [Google Scholar] [CrossRef]
- Wang, X.; Liu, L.; Montagna, C.; Ried, T.; Deng, C.X. Haploinsufficiency of PARP1 accelerates BRCA1-associated centrosome amplification, telomere shortening, genetic instability, apoptosis, and embryonic lethality. Cell Death Differ. 2007, 14, 924–931. [Google Scholar]
- Sugo, N.; Niimi, N.; Aratani, Y.; Masutani, M.; Suzuki, H.; Koyama, H. Decreased PARP-1 levels accelerate embryonic lethality but attenuate neuronal apoptosis in DNA polymerase beta-deficient mice. Biochem. Biophys. Res. Commun. 2007, 354, 656–661. [Google Scholar] [CrossRef]
- Henning, S.M.; Swendseid, M.E.; Coulson, W.F. Male rats fed methyl- and folate-deficient diets with or without niacin develop hepatic carcinomas associated with decreased tissue nad concentrations and altered poly(ADP-ribose) polymerase activity. J. Nutr. 1997, 127, 30–36. [Google Scholar]
- Khodyreva, S.N.; Prasad, R.; Ilina, E.S.; Sukhanova, M.V.; Kutuzov, M.M.; Liu, Y.; Hou, E.W.; Wilson, S.H.; Lavrik, O.I. Apurinic/apyrimidinic (AP) site recognition by the 5'-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. USA 2010, 107, 22090–22095. [Google Scholar]
- Hegde, M.L.; Hazra, T.K.; Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008, 18, 27–47. [Google Scholar] [CrossRef]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar]
- Gagne, J.P.; Isabelle, M.; Lo, K.S.; Bourassa, S.; Hendzel, M.J.; Dawson, V.L.; Dawson, T.M.; Poirier, G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008, 36, 6959–6976. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 2012, 12, 104–120. [Google Scholar]
- Dantzer, F.; de La Rubia, G.; Menissier-De Murcia, J.; Hostomsky, Z.; de Murcia, G.; Schreiber, V. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 2000, 39, 7559–7569. [Google Scholar]
- Friedberg, E.C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 2001, 1, 22–33. [Google Scholar] [CrossRef]
- Ambrose, H.E.; Willimott, S.; Beswick, R.W.; Dantzer, F.; de Murcia, J.M.; Yelamos, J.; Wagner, S.D. Poly(ADP-ribose) polymerase-1 (PARP-1)-deficient mice demonstrate abnormal antibody responses. Immunology 2009, 127, 178–186. [Google Scholar] [CrossRef]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef]
- Paddock, M.N.; Bauman, A.T.; Higdon, R.; Kolker, E.; Takeda, S.; Scharenberg, A.M. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair (Amst) 2011, 10, 338–343. [Google Scholar]
- Mansour, W.Y.; Rhein, T.; Dahm-Daphi, J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010, 38, 6065–6077. [Google Scholar] [CrossRef]
- Hochegger, H.; Dejsuphong, D.; Fukushima, T.; Morrison, C.; Sonoda, E.; Schreiber, V.; Zhao, G.Y.; Saberi, A.; Masutani, M.; Adachi, N., et al. PARP-1 protects homologous recombination from interference by ku and ligase iv in vertebrate cells. EMBO J. 2006, 25, 1305–1314. [Google Scholar] [CrossRef]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef]
- Zhou, B.B.; Bartek, J. Targeting the checkpoint kinases: Chemosensitization versus chemoprotection. Nat. Rev. Cancer 2004, 4, 216–225. [Google Scholar] [CrossRef]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Lobrich, M.; Jeggo, P.A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 2007, 7, 861–869. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar]
- Nam, E.A.; Cortez, D. ATR signalling: More than meeting at the fork. Biochem. J 2011, 436, 527–536. [Google Scholar] [CrossRef]
- Lieber, M.R.; Wilson, T.E. Snapshot: Nonhomologous DNA end joining (NHEJ). Cell 2010, 142. 496–496 e491. [Google Scholar]
- Ruscetti, T.; Lehnert, B.E.; Halbrook, J.; Le Trong, H.; Hoekstra, M.F.; Chen, D.J.; Peterson, S.R. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 1998, 273, 14461–14467. [Google Scholar]
- Alvarez-Gonzalez, R.; Spring, H.; Muller, M.; Burkle, A. Selective loss of poly(ADP-ribose) and the 85-kDa fragment of poly(ADP-ribose) polymerase in nucleoli during alkylation-induced apoptosis of hela cells. J. Biol. Chem. 1999, 274, 32122–32126. [Google Scholar] [CrossRef]
- Soldani, C.; Lazze, M.C.; Bottone, M.G.; Tognon, G.; Biggiogera, M.; Pellicciari, C.E.; Scovassi, A.I. Poly(ADP-ribose) polymerase cleavage during apoptosis: When and where? Exp. Cell Res. 2001, 269, 193–201. [Google Scholar] [CrossRef]
- Beneke, S. Poly(ADP-ribose) polymerase activity in different pathologies--the link to inflammation and infarction. Exp. Gerontol. 2008, 43, 605–614. [Google Scholar] [CrossRef]
- Yu, S.W.; Wang, H.; Poitras, M.F.; Coombs, C.; Bowers, W.J.; Federoff, H.J.; Poirier, G.G.; Dawson, T.M.; Dawson, V.L. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 2002, 297, 259–263. [Google Scholar]
- Tulin, A.; Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at drosophila puff loci. Science 2003, 299, 560–562. [Google Scholar] [CrossRef]
- Simbulan-Rosenthal, C.M.; Ly, D.H.; Rosenthal, D.S.; Konopka, G.; Luo, R.; Wang, Z.Q.; Schultz, P.G.; Smulson, M.E. Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc. Natl. Acad. Sci. USA 2000, 97, 11274–11279. [Google Scholar]
- Frizzell, K.M.; Gamble, M.J.; Berrocal, J.G.; Zhang, T.; Krishnakumar, R.; Cen, Y.; Sauve, A.A.; Kraus, W.L. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J. Biol. Chem. 2009, 284, 33926–33938. [Google Scholar]
- Ogino, H.; Nozaki, T.; Gunji, A.; Maeda, M.; Suzuki, H.; Ohta, T.; Murakami, Y.; Nakagama, H.; Sugimura, T.; Masutani, M. Loss of PARP-1 affects gene expression profile in a genome-wide manner in es cells and liver cells. BMC Genomics 2007, 8, 41. [Google Scholar] [CrossRef]
- Butler, A.J.; Ordahl, C.P. Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol. Cell Biol. 1999, 19, 296–306. [Google Scholar]
- Plaza, S.; Aumercier, M.; Bailly, M.; Dozier, C.; Saule, S. Involvement of poly (ADP-ribose)-polymerase in the Pax-6 gene regulation in neuroretina. Oncogene 1999, 18, 1041–1051. [Google Scholar] [CrossRef]
- Amiri, K.I.; Ha, H.C.; Smulson, M.E.; Richmond, A. Differential regulation of CXC ligand 1 transcription in melanoma cell lines by poly(ADP-ribose) polymerase-1. Oncogene 2006, 25, 7714–7722. [Google Scholar] [CrossRef]
- Chu, S.; Xu, H.; Ferro, T.J.; Rivera, P.X. Poly(ADP-ribose) polymerase-1 regulates vimentin expression in lung cancer cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 293, L1127–1134. [Google Scholar] [CrossRef]
- Maruyama, T.; Nara, K.; Yoshikawa, H.; Suzuki, N. Txk, a member of the non-receptor tyrosine kinase of the tec family, forms a complex with poly(ADP-ribose) polymerase 1 and elongation factor 1alpha and regulates interferon-gamma gene transcription in Th1 cell. Clin. Exp. Immunol. 2007, 147, 164–175. [Google Scholar]
- Wang, J.; Bian, C.; Li, J.; Couch, F.J.; Wu, K.; Zhao, R.C. Poly(ADP-ribose) polymerase-1 down-regulates BRCA2 expression through the brca2 promoter. J. Biol. Chem. 2008, 283, 36249–36256. [Google Scholar]
- Ono, T.; Kaneda, T.; Muto, A.; Yoshida, T. Positive transcriptional regulation of the human micro opioid receptor gene by poly(ADP-ribose) polymerase-1 and increase of its DNA binding affinity based on polymorphism of G-172 ->T. J. Biol. Chem. 2009, 284, 20175–20183. [Google Scholar]
- Reinemund, J.; Seidel, K.; Steckelings, U.M.; Zaade, D.; Klare, S.; Rompe, F.; Katerbaum, M.; Schacherl, J.; Li, Y.; Menk, M., et al. Poly(ADP-ribose) polymerase-1 (parp-1) transcriptionally regulates angiotensin AT2 receptor (AT2R) and AT2R binding protein (ATBP) genes. Biochem. Pharmacol. 2009, 77, 1795–1805. [Google Scholar]
- Kang, X.; Kim, H.J.; Ramirez, M.; Salameh, S.; Ma, X. The septic shock-associated Il-10 -1082 A > G polymorphism mediates allele-specific transcription via poly(ADP-ribose) polymerase 1 in macrophages engulfing apoptotic cells. J. Immunol. 2010, 184, 3718–3724. [Google Scholar] [CrossRef]
- Zaniolo, K.; Desnoyers, S.; Leclerc, S.; Guerin, S.L. Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of SP1: A nuclear target protein of PARP-1. BMC Mol. Biol. 2007, 8, 96. [Google Scholar]
- Huang, D.; Yang, C.; Wang, Y.; Liao, Y.; Huang, K. PARP-1 suppresses adiponectin expression through poly(ADP-ribosyl)ation of PPAR gamma in cardiac fibroblasts. Cardiovasc. Res. 2009, 81, 98–107. [Google Scholar] [CrossRef]
- Lonn, P.; van der Heide, L.P.; Dahl, M.; Hellman, U.; Heldin, C.H.; Moustakas, A. PARP-1 attenuates Smad-mediated transcription. Mol. Cell 2010, 40, 521–532. [Google Scholar] [CrossRef]
- Gao, F.; Kwon, S.W.; Zhao, Y.; Jin, Y. Parp1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation. J. Biol. Chem. 2009, 284, 22263–22273. [Google Scholar] [CrossRef]
- Hassa, P.O.; Covic, M.; Hasan, S.; Imhof, R.; Hottiger, M.O. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J. Biol. Chem. 2001, 276, 45588–45597. [Google Scholar]
- Hassa, P.O.; Hottiger, M.O. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell. Mol. Life. Sci. 2002, 59, 1534–1553. [Google Scholar] [CrossRef]
- Erener, S.; Petrilli, V.; Kassner, I.; Minotti, R.; Castillo, R.; Santoro, R.; Hassa, P.O.; Tschopp, J.; Hottiger, M.O. Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-kappaB target genes. Mol. Cell 2012, 46, 200–211. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef]
- Thorslund, T.; West, S.C. BRCA2: A universal recombinase regulator. Oncogene 2007, 26, 7720–7730. [Google Scholar] [CrossRef]
- Jensen, R.B.; Carreira, A.; Kowalczykowski, S.C. Purified human BRCA2 stimulates Rad51-mediated recombination. Nature 2010, 467, 678–683. [Google Scholar]
- Huang, J.; Gong, Z.; Ghosal, G.; Chen, J. SOSS complexes participate in the maintenance of genomic stability. Mol. Cell 2009, 35, 384–393. [Google Scholar] [CrossRef]
- Navadgi-Patil, V.M.; Burgers, P.M. A tale of two tails: Activation of DNA damage checkpoint kinase Mec1/ATR by the 9-1-1 clamp and by Dpb11/TopBP1. DNA Repair (Amst) 2009, 8, 996–1003. [Google Scholar]
- De Piccoli, G.; Torres-Rosell, J.; Aragon, L. The unnamed complex: What do we know about SMC5-SMC6? Chromosome Res. 2009, 17, 251–263. [Google Scholar] [CrossRef]
- Fekairi, S.; Scaglione, S.; Chahwan, C.; Taylor, E.R.; Tissier, A.; Coulon, S.; Dong, M.Q.; Ruse, C.; Yates, J.R., 3rd; Russell, P., et al. Human SLX4 is a holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 2009, 138, 78–89. [Google Scholar]
- Svendsen, J.M.; Smogorzewska, A.; Sowa, M.E.; O'Connell, B.C.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. Mammalian BTBD12/SLX4 assembles a holliday junction resolvase and is required for DNA repair. Cell 2009, 138, 63–77. [Google Scholar] [CrossRef]
- Li, W.; Ma, H. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 2006, 16, 402–412. [Google Scholar] [CrossRef]
- Regnell, C.E.; H., G.A.; Sejersted, Y.; Medin, T.; Moldestad, O.; Rolseth, V.; Krokeide, S. Z.; Suganthan, R.; Luna, L.; Bjoras, M.; Bergersen, L.H. Hippocampal adult neurogenesis is maintained by NEIL3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Reports 2012. [Google Scholar] [CrossRef]
- Zhu, J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef]
- Bellacosa, A. Role of MED1 (MBD4) gene in DNA repair and human cancer. J. Cell Physiol. 2001, 187, 137–144. [Google Scholar] [CrossRef]
- Nishi, R.; Okuda, Y.; Watanabe, E.; Mori, T.; Iwai, S.; Masutani, C.; Sugasawa, K.; Hanaoka, F. Centrin 2 stimulates nucleotide excision repair by interacting with Xeroderma Pigmentosum group C protein. Mol. Cell. Biol. 2005, 25, 5664–5674. [Google Scholar] [CrossRef]
- Aas, P.A.; Otterlei, M.; Falnes, P.O.; Vagbo, C.B.; Skorpen, F.; Akbari, M.; Sundheim, O.; Bjoras, M.; Slupphaug, G.; Seeberg, E., et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar]
- Duncan, T.; Trewick, S.C.; Koivisto, P.; Bates, P.A.; Lindahl, T.; Sedgwick, B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. USA 2002, 99, 16660–16665. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef]
- Chou, D.M.; Adamson, B.; Dephoure, N.E.; Tan, X.; Nottke, A.C.; Hurov, K.E.; Gygi, S.P.; Colaiacovo, M.P.; Elledge, S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 2010, 107, 18475–18480. [Google Scholar]
- Asher, G.; Reinke, H.; Altmeyer, M.; Gutierrez-Arcelus, M.; Hottiger, M.O.; Schibler, U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 2010, 142, 943–953. [Google Scholar] [CrossRef]
- Vidakovic, M.; Gluch, A.; Qiao, J.; Oumard, A.; Frisch, M.; Poznanovic, G.; Bode, J. PARP-1 expression in the mouse is controlled by an autoregulatory loop: PARP-1 binding to an upstream S/MAR element and to a novel recognition motif in its promoter suppresses transcription. J. Mol. Biol. 2009, 388, 730–750. [Google Scholar] [CrossRef]
- Xu, X.L.; Xing, B.C.; Han, H.B.; Zhao, W.; Hu, M.H.; Xu, Z.L.; Li, J.Y.; Xie, Y.; Gu, J.; Wang, Y., et al. The properties of tumor-initiating cells from a hepatocellular carcinoma patient's primary and recurrent tumor. Carcinogenesis 2009, 31, 167–174. [Google Scholar]
- Shimizu, S.; Nomura, F.; Tomonaga, T.; Sunaga, M.; Noda, M.; Ebara, M.; Saisho, H. Expression of poly(ADP-ribose) polymerase in human hepatocellular carcinoma and analysis of biopsy specimens obtained under sonographic guidance. Oncol. Rep. 2004, 12, 821–825. [Google Scholar]
- Goldberg, M. S.; Xing, D.; Ren, Y.; Orsulic, S.; Bhatia, S. N.; Sharp, P. A. Nanoparticle-mediated delivery of siRNA targeting Parp1 extends survival of mice bearing tumors derived from Brca1-deficient ovarian cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 745–750. [Google Scholar]
- Mabley, J.G.; Horvath, E.M.; Murthy, K.G.; Zsengeller, Z.; Vaslin, A.; Benko, R.; Kollai, M.; Szabo, C. Gender differences in the endotoxin-induced inflammatory and vascular responses: Potential role of poly(ADP-ribose) polymerase activation. J. Pharmacol. Exp. Ther. 2005, 315, 812–820. [Google Scholar] [CrossRef]
- Liu, T.T.; Fang, Y.; Xiong, H.; Chen, T.Y.; Ni, Z.P.; Luo, J.F.; Zhao, N.Q.; Shen, X.Z. A case-control study of the relationship between hepatitis B virus DNA level and risk of hepatocellular carcinoma in qidong, china. World J. Gastroenterol. 2008, 14, 3059–3063. [Google Scholar]
- Chan, H.L.; Wong, V.W.; Wong, G.L.; Chim, A.M.; Lai, L.H.; Sung, J.J. Evaluation of impact of serial hepatitis B virus DNA levels on development of hepatocellular carcinoma. J. Clin. Microbiol. 2009, 47, 1830–1836. [Google Scholar] [CrossRef]
- Kwon, J.H.; Choi, J.Y.; Jang, J.W.; Bae, S.H.; Yoon, S.K.; Yang, J.M.; Han, N.I.; Lee, C.D.; Lee, Y.S.; Chung, K.W. Impact of serial hepatitis B virus DNA on hepatocellular carcinoma development in patients with liver cirrhosis. Intervirology 2009, 53, 111–118. [Google Scholar]
- Hagen, T.M.; Huang, S.; Curnutte, J.; Fowler, P.; Martinez, V.; Wehr, C.M.; Ames, B.N.; Chisari, F.V. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 1994, 91, 12808–12812. [Google Scholar]
- Ohsaki, E.; Ueda, K.; Sakakibara, S.; Do, E.; Yada, K.; Yamanishi, K. Poly(ADP-ribose) polymerase 1 binds to kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 2004, 78, 9936–9946. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Tang, Q.; Maul, G.G.; Yuan, Y. Kaposi's sarcoma-associated herpesvirus ori-lyt-dependent DNA replication: Involvement of host cellular factors. J. Virol. 2008, 82, 2867–2882. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ko, H.L.; Ren, E.C. Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules 2012, 2, 524-548. https://doi.org/10.3390/biom2040524
Ko HL, Ren EC. Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules. 2012; 2(4):524-548. https://doi.org/10.3390/biom2040524
Chicago/Turabian StyleKo, Hui Ling, and Ee Chee Ren. 2012. "Functional Aspects of PARP1 in DNA Repair and Transcription" Biomolecules 2, no. 4: 524-548. https://doi.org/10.3390/biom2040524