Hyaluronidases Have Strong Hydrolytic Activity toward Chondroitin 4-Sulfate Comparable to that for Hyaluronan
Abstract
:1. Introduction
2. Results
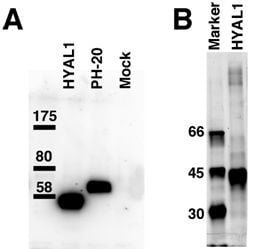
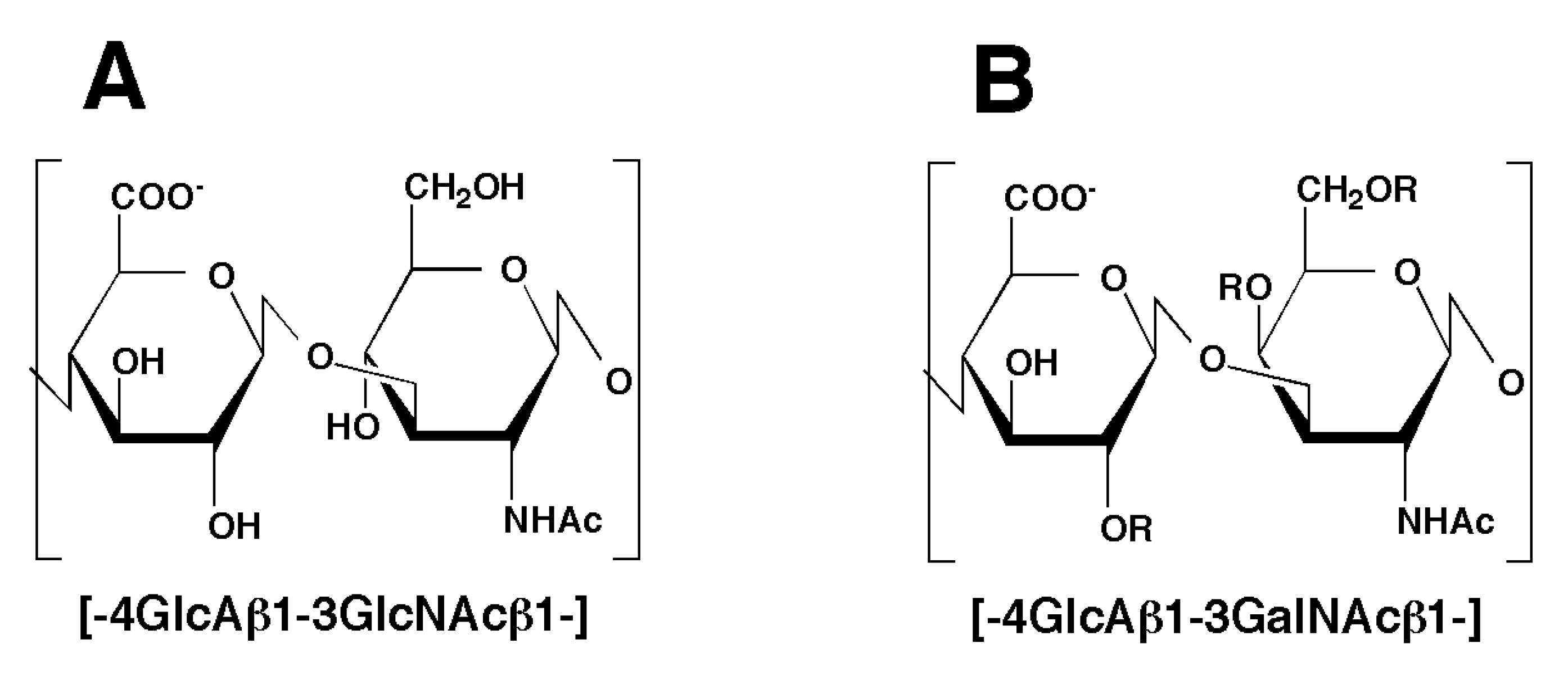
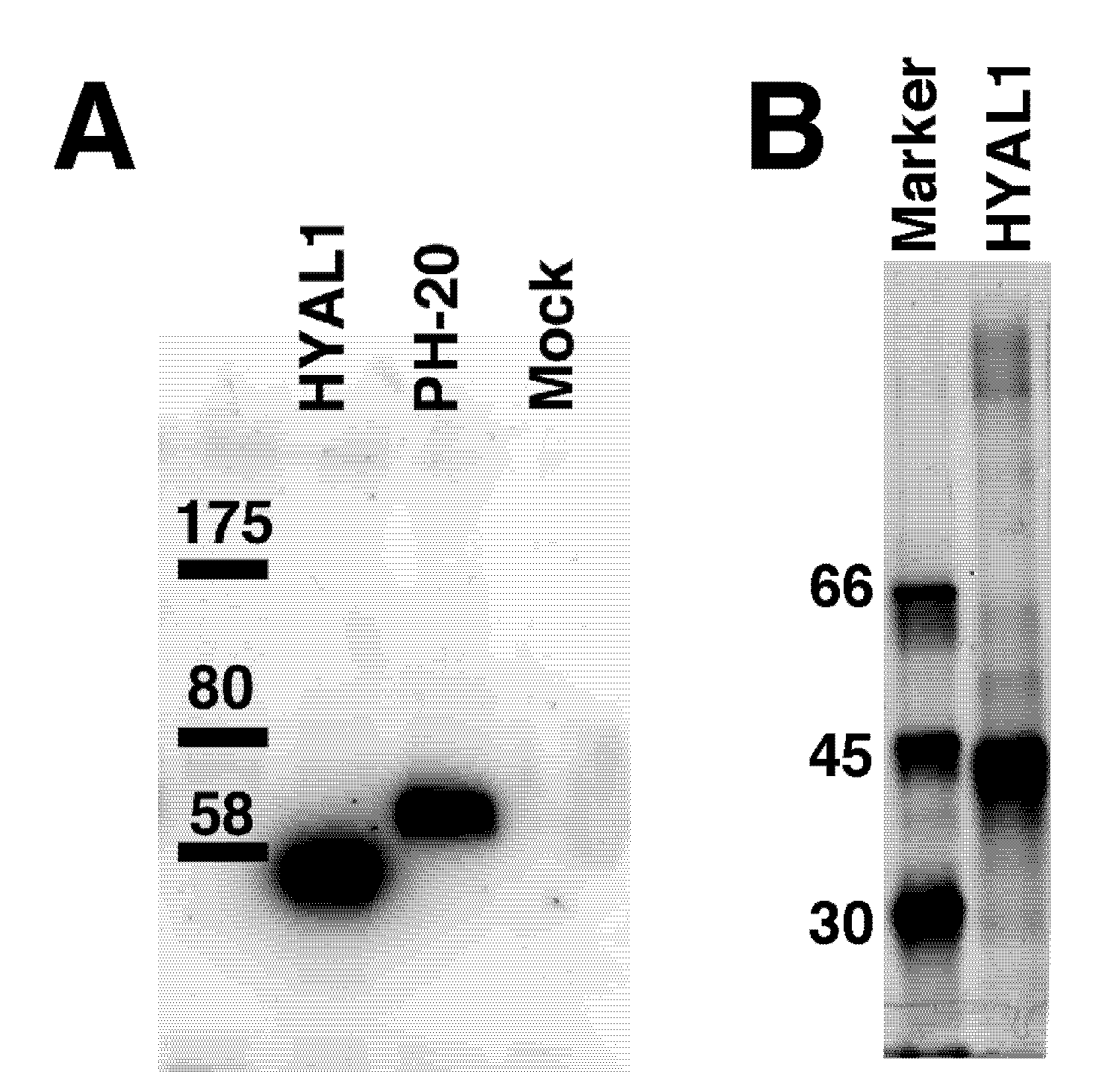
2.1. Hydrolytic Activity of HYAL1 and SPAM1 toward CS Variants as well as HA
| Substrate | Apparent Km | Apparent Vmax | Apparent Vmax/Km |
|---|---|---|---|
| mM as disaccharides | pmol/min | ||
| HA | 0.40 | 23.0 | 57.5 |
| CS-A | 0.21 | 30.5 | 145 |
| CS-C | 0.64 | 10.8 | 16.9 |
| Chn | 1.20 | 9.0 | 7.5 |
| Substrate | Apparent Km | Apparent Vmax | Apparent Vmax/Km |
|---|---|---|---|
| mM as disaccharides | pmol/min | ||
| HA | 0.20 | 0.92 | 4.5 |
| CS-A | 0.13 | 0.94 | 7.5 |
| CS-C | 0.48 | 0.28 | 0.6 |
| Chn | 1.02 | 1.55 | 1.5 |
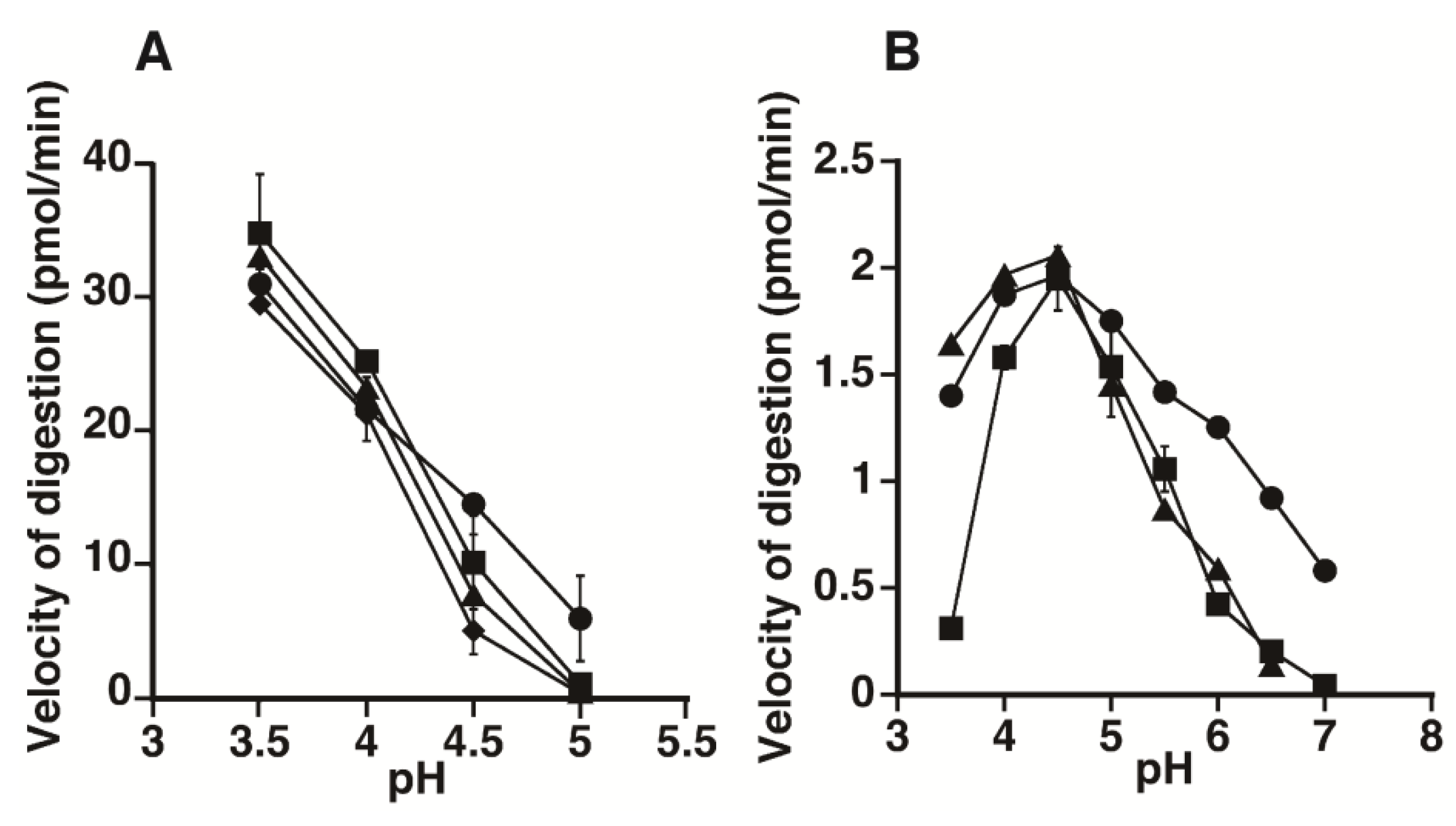
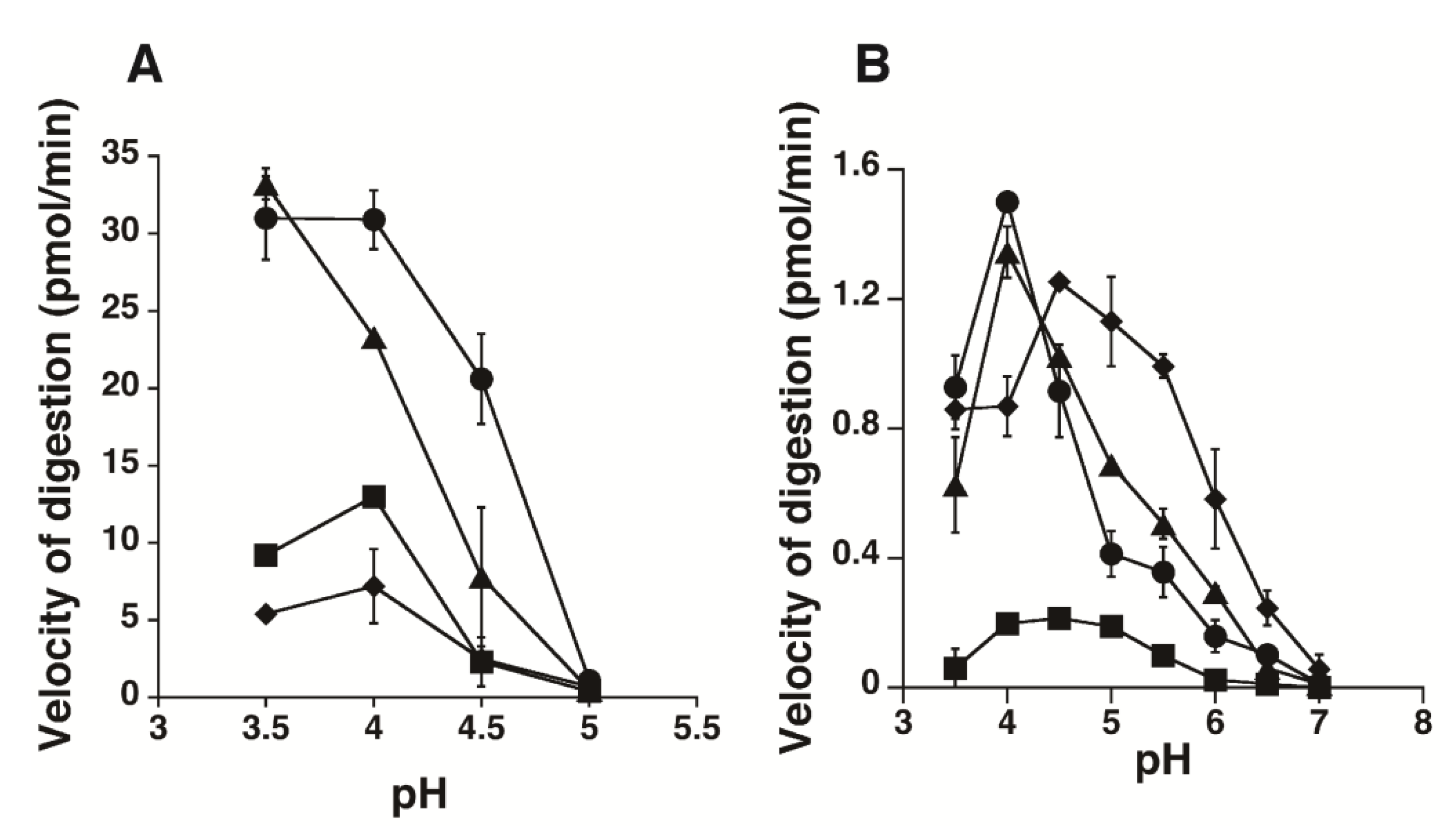
2.2. Kinetic Analysis of CS-Degrading Activity of HYAL1 and SPAM1
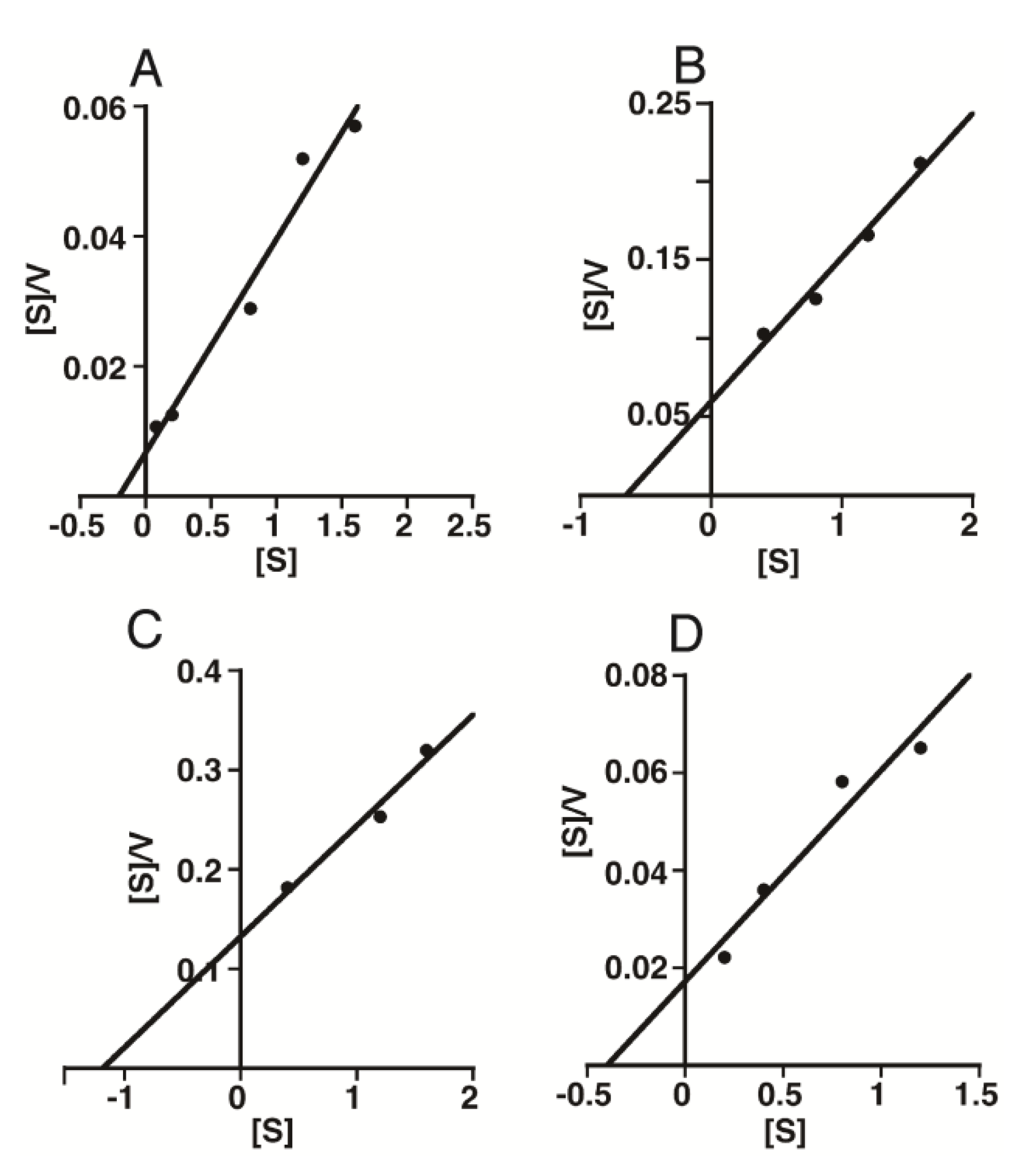
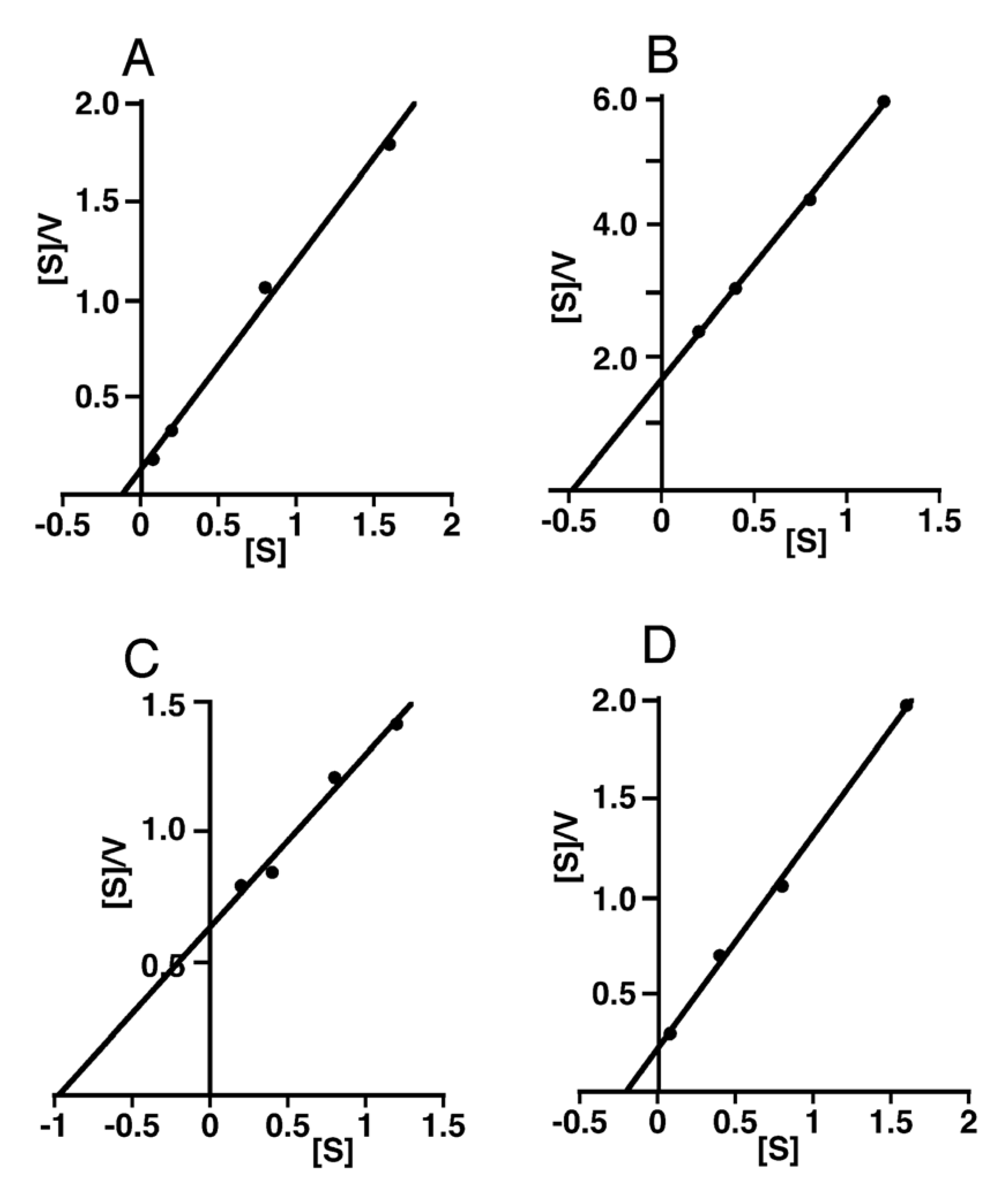
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Cloning of SPAM1 cDNA
4.3. Construction of an Expression Vector Containing a cDNA Fragment Encoding a Soluble Form of HYAL1 or SPAM1
4.4. Expression of a Soluble Form of HYAL1 or SPAM1
4.5. Measurement of the Enzymatic Activity
4.6. Western Blotting
5. Conclusions
Abbreviations
| 2AB | 2-aminobenzamide |
| Chn | chondroitin |
| CS | chondroitin sulfate |
| HA | hyaluronan |
| HPLC | high performance liquid chromatography |
Acknowledgments
Conflict of Interest
References
- Sugahara, K.; Yamada, S. Structure and function of oversulfated chondroitin sulfate variants: Unique sulfation patterns and neuroregulatory activities. Trends Glycosci. Glycotechnol. 2000, 12, 321–349. [Google Scholar] [CrossRef]
- Sugahara, K.; Mikami, T.; Uyama, T.; Mizuguchi, S.; Nomura, K.; Kitagawa, H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003, 13, 612–620. [Google Scholar] [CrossRef]
- Rauch, U.; Kappler, L. Chondroitin/dermatan sulfates in the central nervous system: Their structures and functions in health and disease. Adv. Pharmacol. 2006, 53, 337–356. [Google Scholar] [CrossRef]
- Sugahara, K.; Mikami, T. Chondroitin/dermatan sulfate in the central nervous system. Curr. Opin. Struct. Biol. 2007, 17, 536–545. [Google Scholar]
- Uyama, T.; Kitagawa, H.; Sugahara, K. Biosynthesis of glycosaminoglycans and proteoglycans. In Comprehensive Glycoscience; Kamerling, J.P., Ed.; Elsevier: Amsterdam, the Netherlands, 2007; Volume 3, pp. 79–104. [Google Scholar]
- Prabhakar, V.; Sasisekharan, R. The biosynthesis and catabolism of galactosaminoglycans. Adv. Pharmacol. 2006, 53, 69–115. [Google Scholar]
- Kresse, H.; Glössl, J. Glycosaminoglycan degradation. Adv. Enzymol. Relat. Areas Mol. Biol. 1987, 60, 217–311. [Google Scholar]
- Csoka, A.B.; Frost, G.I.; Stern, R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001, 20, 499–508. [Google Scholar]
- Jedrzejas, M.J.; Stern, R. Structures of vertebrate hyaluronidases and their unique enzymatic mechanism of hydrolysis. Proteins 2005, 61, 227–238. [Google Scholar] [CrossRef]
- Kaneiwa, T.; Mizumoto, S.; Sugahara, K.; Yamada, S. Identification of human hyaluronidase-4 as a novel chondroitin sulfate hydrolase that preferentially cleaves the galactosaminidic linkage in the trisulfated tetrasaccharide sequence. Glycobiology 2010, 20, 300–309. [Google Scholar] [CrossRef]
- Yamada, S. Chondroitin sulfate-specific novel hydrolase in human. Adv. Exp. Med. Biol. 2012, 749, 47–56. [Google Scholar]
- Csoka, A.B.; Scherer, S.W.; Stern, R. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics 1999, 60, 356–361. [Google Scholar] [CrossRef]
- Kaneiwa, T.; Miyazaki, A.; Kogawa, R.; Mizumoto, S.; Sugahara, K.; Yamada, S. Identification of amino acid residues required for the substrate specificity of human and mouse chondroitin sulfate hydrolase (conventional hyaluronidase-4). J. Biol. Chem. 2012, in press. [Google Scholar]
- Sugahara, K.; Tanaka, Y.; Yamada, S. Preparation of a series of sulfated tetrasaccharides from shark cartilage chondroitin sulfate D using testicular hyaluronidase and structure determination by 500 MHz 1H NMR spectroscopy. Glycoconj. J. 1996, 13, 609–619. [Google Scholar] [CrossRef]
- Sugahara, K.; Tanaka, Y.; Yamada, S.; Seno, N.; Kitagawa, H.; Haslam, S.M.; Morris, HR.; Dell, A. Novel sulfated oligosaccharides containing 3-O-sulfated glucuronic acid from king crab cartilage chondroitin sulfate K. Unexpected degradation by chondroitinase ABC. J. Biol. Chem. 1996, 271, 26745–26754. [Google Scholar]
- Guntenhöner, M.W.; Pogrel, M.A.; Stern, R. A substrate-gel assay for hyaluronidase activity. Matrix 1992, 12, 388–396. [Google Scholar]
- Di Ferrante, N. Turbidimetric measurement of acid mucopolysaccharides and hyaluronidase activity. J. Biol. Chem. 1956, 220, 303–306. [Google Scholar]
- Tung, J.S.; Mark, G.E.; Hollis, G.F. A microplate assay for hyaluronidase and hyaluronidase inhibitors. Anal. Biochem. 1994, 223, 149–152. [Google Scholar]
- Stern, M. An ELISA-like assay for hyaluronidase and hyaluronidase inhibitors. Matrix 1992, 12, 397–403. [Google Scholar] [CrossRef]
- Müllegger, J.; Reitinger, S.; Lepperdinger, G. Hapten-labeled hyaluronan, a substrate to monitor hyaluronidase activity by enhanced chemiuminescence-assisted detection on filter blots. Anal. Biochem. 2001, 293, 291–293. [Google Scholar]
- Bonner, W., Jr.; Cantey, E.Y. Colorimetric method for determination of serum hyaluronidase activity. Clin. Chim. Acta 1966, 13, 746–752. [Google Scholar] [CrossRef]
- Baba, D.; Kashiwabara, S.; Honda, A.; Yamagata, K.; Wu, Q.; Ikawa, M.; Okabe, M.; Baba, T. Mouse sperm lacking cell surface hyaluronidase PH-20 can pass through the layer of cumulus cells and fertilize the egg. J. Biol. Chem. 2002, 277, 30310–30314. [Google Scholar]
- Lin, Y.; Mahan, K.; Lathrop, W.F.; Myles, D.G.; Primakoff, P. A hyaluronidase activity of the sperm plasma membrane protein PH-20 enables sperm to penetrate the cumulus cell layer surrounding the egg. J. Cell. Biol. 1994, 125, 1157–1163. [Google Scholar]
- Kaneiwa, T.; Yamada, S.; Mizumoto, S.; Motaño, A.M.; Mitani, S.; Sugahara, K. Identification of novel chondroitin hydrolase in Caenorhabditis. elegans. J. Biol. Chem. 2008, 283, 14971–14979. [Google Scholar]
- Yamada, S.; Mizumoto, S.; Sugahara, K. Chondroitin hydrolase in Caenorhabditis. elegans. Trends Glycosci. Glycotechnol. 2009, 21, 149–162. [Google Scholar] [CrossRef]
- Harada, H.; Takahashi, M. CD44-dependent intracellular and extracellular catabolism of hyaluronic acid by hyaluronidase-1 and -2. J. Biol. Chem. 2007, 282, 5597–5607. [Google Scholar] [CrossRef]
- Yamada, S.; Sugahara, K. Potential therapeutic application of chondroitin sulfate/dermatan sulfate. Curr. Drug Discov. Technol. 2008, 5, 289–301. [Google Scholar]
- Ohtake-Niimi, S.; Kondo, S.; Ito, T.; Kakehi, S.; Ohta, T.; Habuchi, H.; Kimata, K.; Habuchi, O. Mice deficient in N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase are unable to synthesize chondroitin/dermatan sulfate containing N-acetylgalactosamine 4,6-bissulfate residues and exhibit decreased protease activity in bone marrow-derived mast cells. J. Biol. Chem. 2010, 285, 20793–20805. [Google Scholar]
- Triggs-Raine, B.; Salo, T.J.; Zhang, H.; Wicklow, B.A.; Natowicz, M.R. Mutations in HYAL1, a member of a tandemly distributed multigene family encoding disparate hyaluronidase activities, cause a newly described lysosomal disorder, mucopolysaccharidosis IX. Proc. Natl. Acad. Sci. USA 1999, 96, 6296–6300. [Google Scholar]
- Yamada, S.; Sugahara, K.; Özbek, S. Evolution of glycosaminoglycans: Comparative biochemical study. Commun. Integr. Biol. 2011, 4, 150–158. [Google Scholar] [CrossRef]
- Yamada, S.; Van Die, I.; Van den Eijnden, D.H.; Yokota, A.; Kitagawa, H.; Sugahara, K. Demonstration of glycosaminoglycans in Caenorhabditis. elegans. FEBS Lett. 1999, 459, 327–331. [Google Scholar] [CrossRef]
- Yamada, S.; Morimoto, H.; Fujisawa, T.; Sugahara, K. Glycosaminoglycans in Hydra magnipapillate (Hydrozoa, Cnidaria): Demonstration of chondroitin in the developing nematocyst, the sting organelle, and structural characterization of glycosaminoglycans. Glycobiology. 2007, 17, 886–894. [Google Scholar] [CrossRef]
- Wessels, M.R.; Moses, A.E.; Goldberg, J.B.; DiCesare, T.J. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc. Natl. Acad. Sci. USA 1991, 88, 8317–8321. [Google Scholar] [CrossRef]
- Volpi, N.; Maccari, F. Purification and characterization of hyaluronic acid from the mollusc bivalve Mytilus. galloprovincialis. Biochimie. 2003, 85, 619–625. [Google Scholar] [CrossRef]
- Kinoshita, A.; Sugahara, K. Microanalysis of glycosaminoglycan-derived oligosaccharides labeled with a fluorophore 2-aminobenzamide by high-performance liquid chromatography: application to disaccharide composition analysis and exosequencing of oligosaccharides. Anal. Biochem. 1999, 269, 367–378. [Google Scholar]
- Kawashima, H.; Atarashi, K.; Hirose, M.; Hirose, J.; Yamada, S.; Sugahara, K.; Miyasaka, M. Oversulfated chondroitin/dermatan sulfates containing GlcAα1/IdoAβ1–3GalNAc(4,6-O-disulfate) interact with L- and P-selectin and chemokines. J. Biol. Chem. 2002, 277, 12921–12930. [Google Scholar]
- Gushulak, L.; Hemming, R.; Martin, D.; Seyrantepe, V.; Pshezhetsky, A.; Triggs-Raine, B. Hyaluronidase 1 and β-exosaminidase have redundant functions in hyaluronan and chondroitin sulfate degradation. J. Biol. Chem. 2012, 287, 16689–16697. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Honda, T.; Kaneiwa, T.; Mizumoto, S.; Sugahara, K.; Yamada, S. Hyaluronidases Have Strong Hydrolytic Activity toward Chondroitin 4-Sulfate Comparable to that for Hyaluronan. Biomolecules 2012, 2, 549-563. https://doi.org/10.3390/biom2040549
Honda T, Kaneiwa T, Mizumoto S, Sugahara K, Yamada S. Hyaluronidases Have Strong Hydrolytic Activity toward Chondroitin 4-Sulfate Comparable to that for Hyaluronan. Biomolecules. 2012; 2(4):549-563. https://doi.org/10.3390/biom2040549
Chicago/Turabian StyleHonda, Tomoko, Tomoyuki Kaneiwa, Shuji Mizumoto, Kazuyuki Sugahara, and Shuhei Yamada. 2012. "Hyaluronidases Have Strong Hydrolytic Activity toward Chondroitin 4-Sulfate Comparable to that for Hyaluronan" Biomolecules 2, no. 4: 549-563. https://doi.org/10.3390/biom2040549