Variation in the Subcellular Localization and Protein Folding Activity among Arabidopsis thaliana Homologs of Protein Disulfide Isomerase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Domain Architecture and Sequence Motifs of Arabidopsis PDIA1 and PDIA6 Homologs
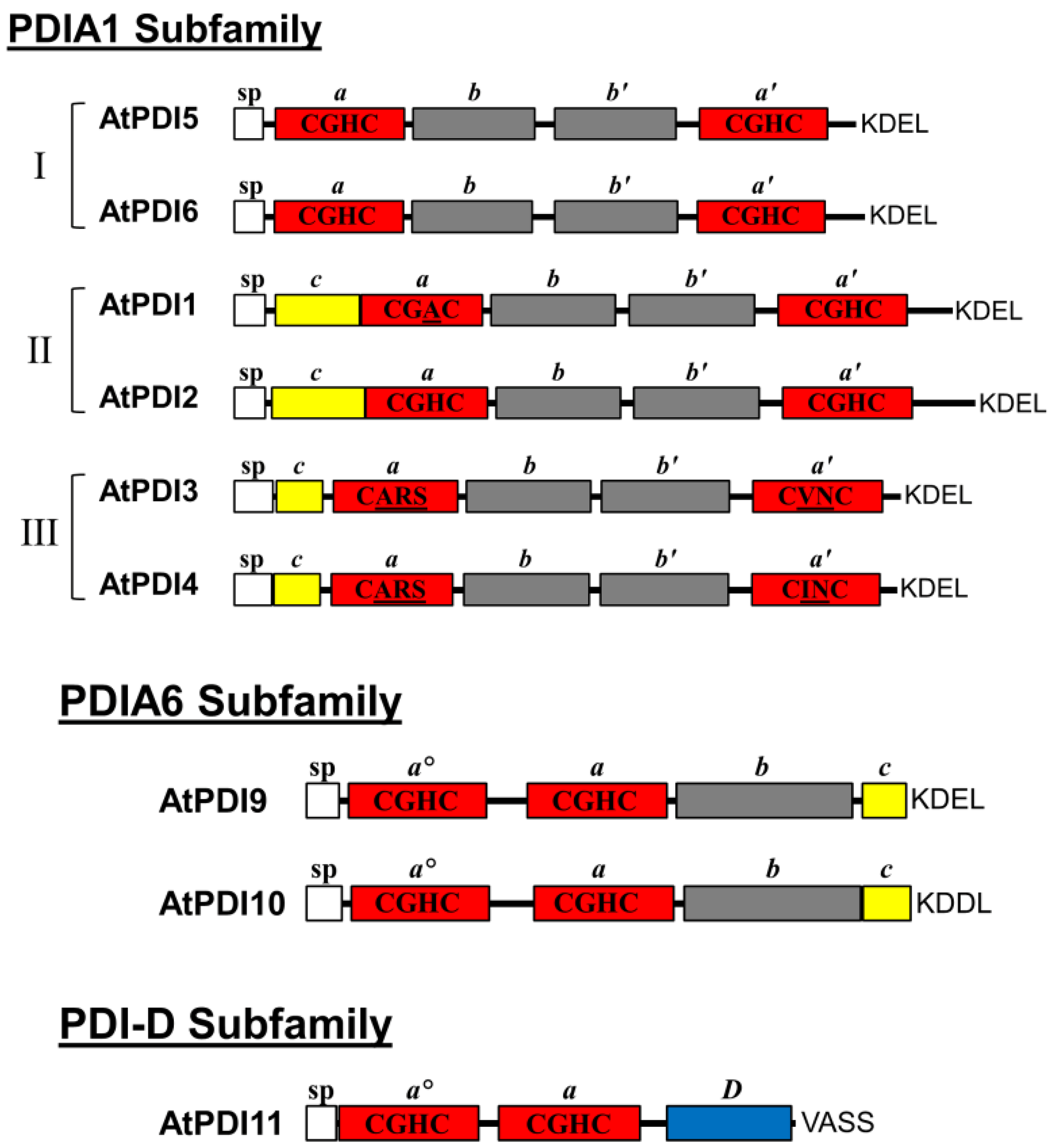
| Name | AGI Number | Domain Arrangement | Active Site Sequence | Conserved Arg Positions | Alternate Names |
|---|---|---|---|---|---|
| PDIA1 | |||||
| Group I | |||||
| PDI5 | At1g21750 | a-b-b′-a′ | CGHC/CGHC | R127/R468 | PDIL1-1/PDIL1-1 |
| PDI6 | At1g77510 | a-b-b′-a′ | CGHC/CGHC | R126/R466 | PDIL1-2/PDIL1-2 |
| Group II | |||||
| PDI1 | At3g54960 | a-b-b′-a′ | CGAC/CGHC | R190/R532 | PDIL1-3/PDIL2-1 |
| PDI2 | At5g60640 | a-b-b′-a′ | CGHC/CGHC | R194/R536 | PDIL1-4/PDIL2-2 |
| Group III | |||||
| PDI3 | At1g52260 | a-b-b′-a′ | CARS/CVNC | (S170)/(L485) | PDIL1-5/PDIL 3-1 |
| PDI4 | At3g16110 | a-b-b′-a′ | CARS/CINC | (F168)/(S510) | PDIL1-6/PDIL3-2 |
| PDIA6 | |||||
| PDI9 | At2g32920 | ao-a-b | CGHC/CGHC | R122/R255 | PDIL2-3/PDIL5-2 |
| PDI10 | At1g04980 | ao-a-b | CGHC/CGHC | R124/R260 | PDIL2-2/PDIL5-1 |
| PDI-D | |||||
| PDI11 | At2g47470 | ao-a-D | CGHC/CGHC | R117/R236 | PDIL2-1/PDIL4-1 |
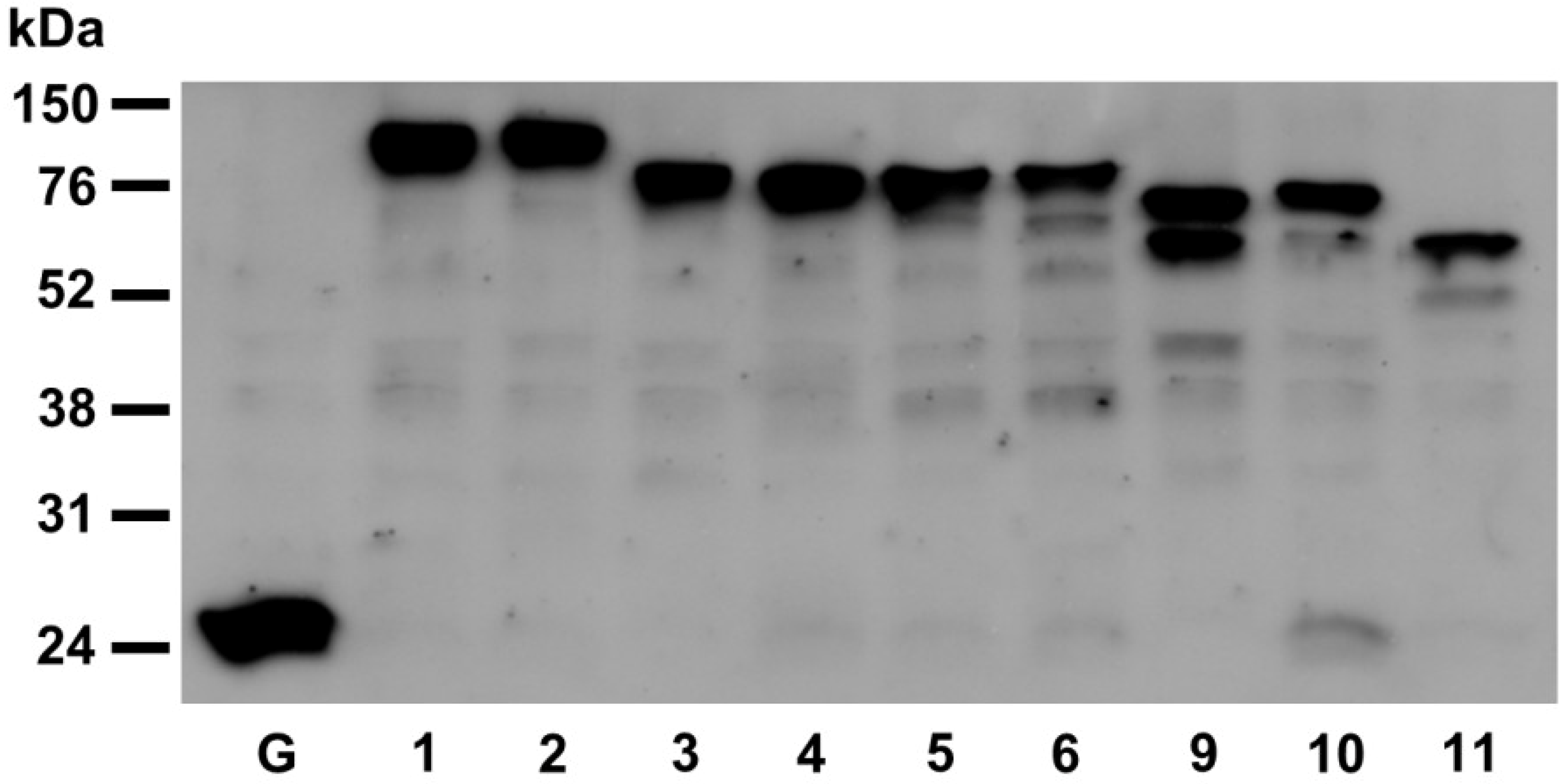
2.2. Fluorescent Protein Fusions of the Arabidopsis PDIA1 and PDIA6 Exhibit Distinct Localization Patterns When Transiently Expressed in Protoplasts
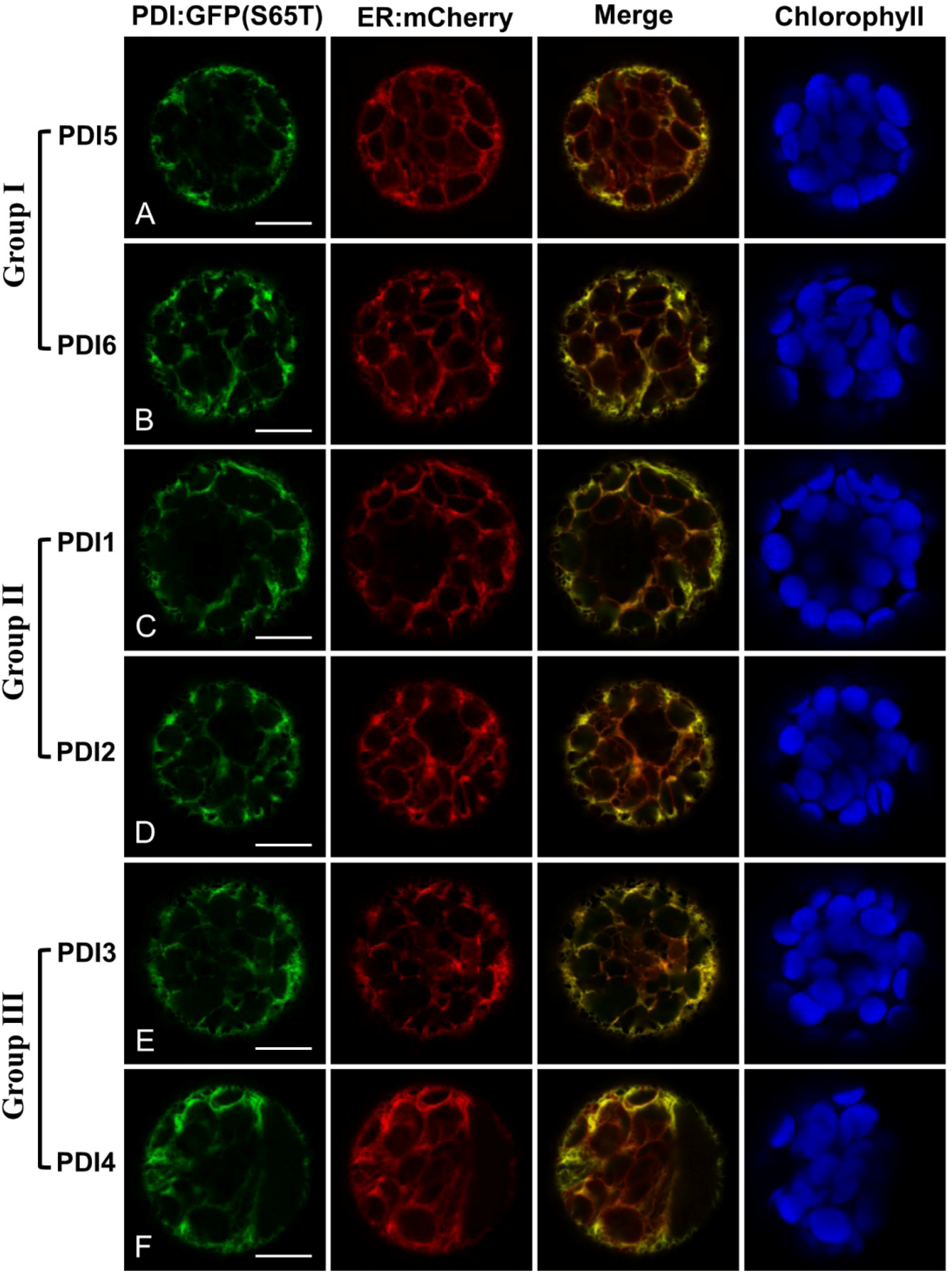
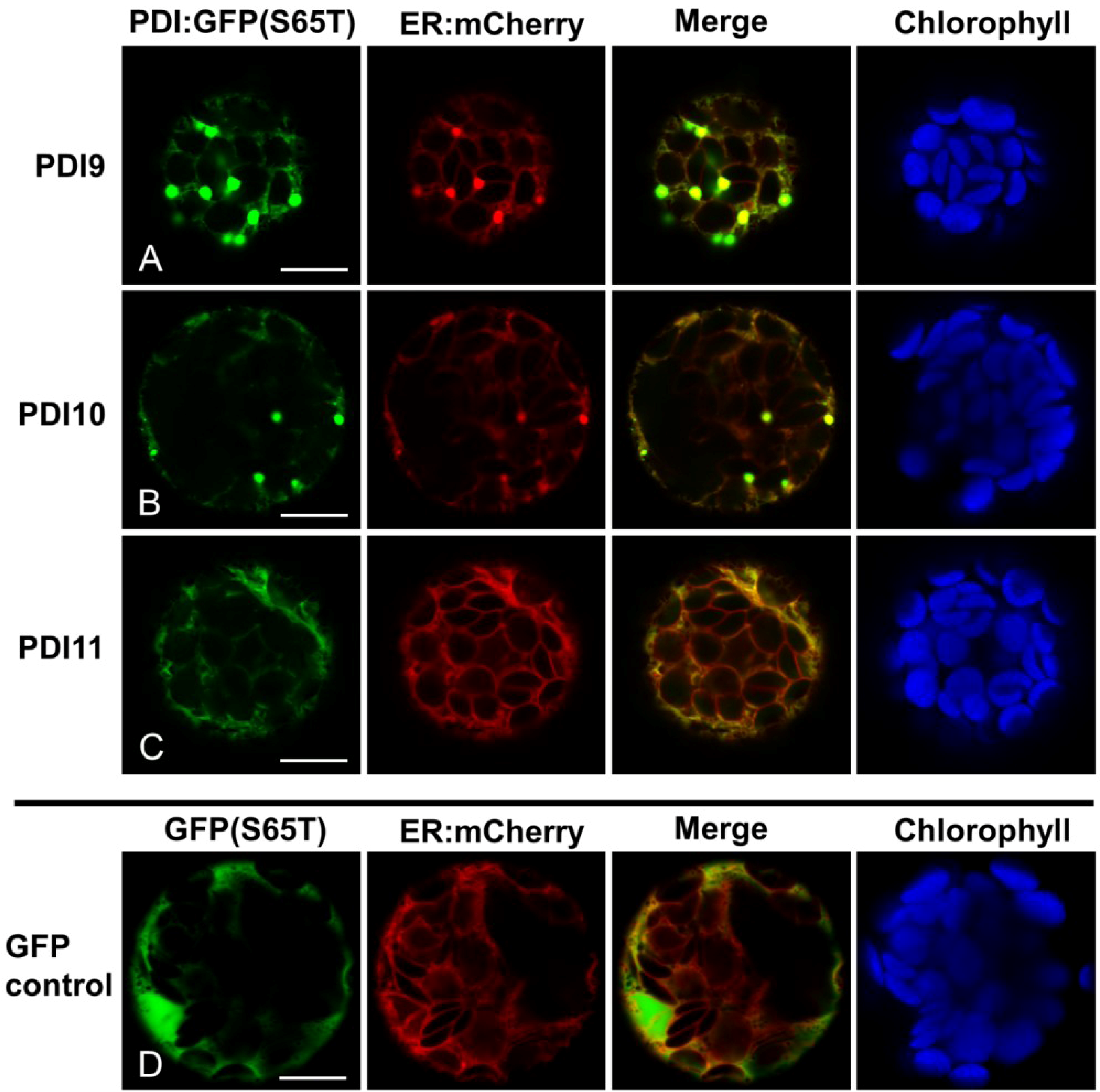

2.3. Restoration of Alkaline Phosphatase Activity in the Escherichia coli Protein Folding Mutant, dsbA−, by a Subset of Arabidopsis PDIs
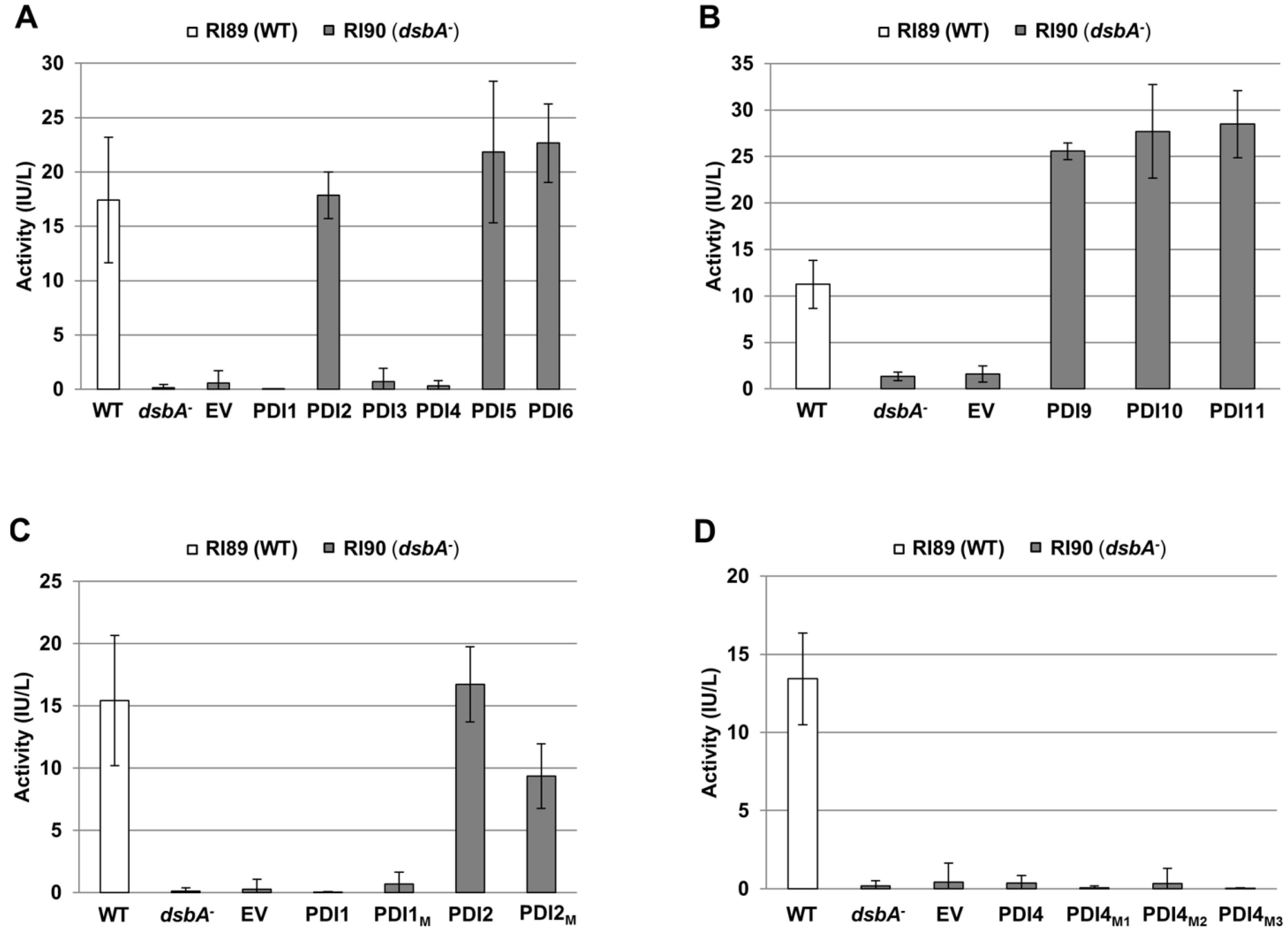
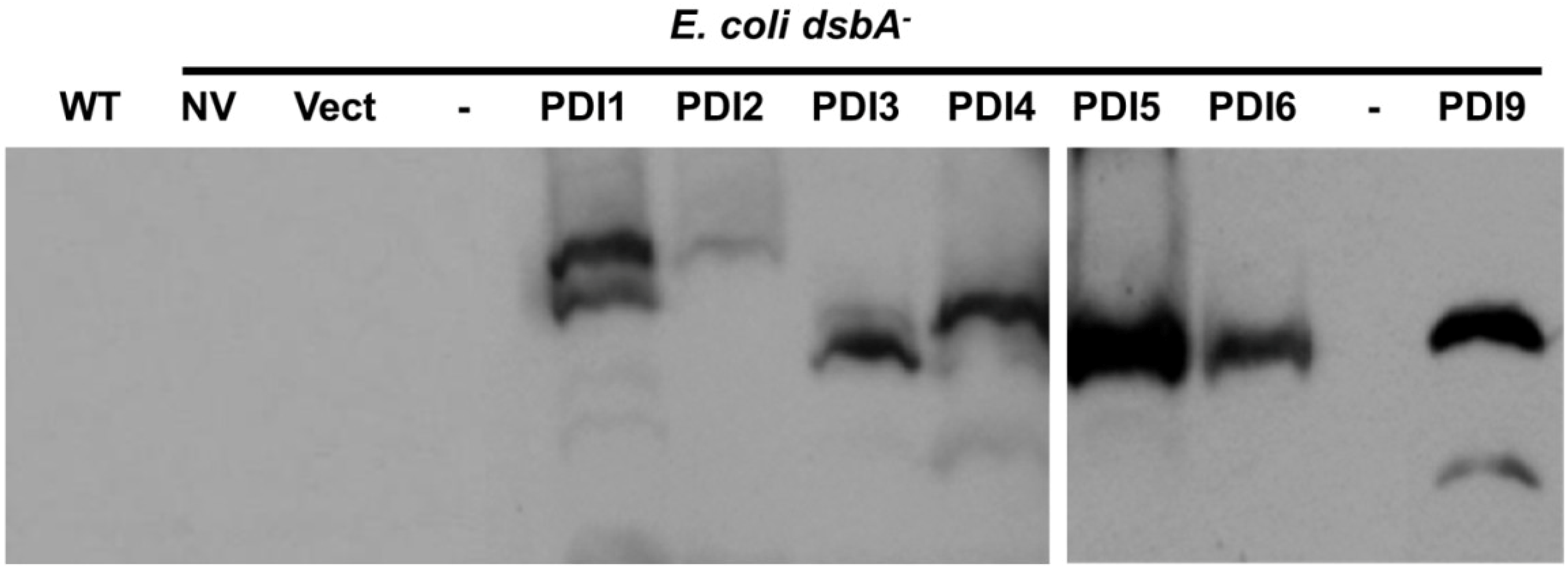
2.4. Functions of PDIA1 and PDIA6 Orthologs in Plants
2.4.1. PDIA1-like Subfamily, Group I (AtPDI5, AtPDI6)
2.4.2. PDIA1-like Subfamily, Group II (AtPDI1, AtPDI2)
2.4.3. PDIA1-like Subfamily, Group III (AtPDI3, AtPDI4)
2.4.4. PDIA6-like Subfamily (AtPDI9, AtPDI10)
3. Experimental
3.1. Constructs
| Construct | PCR Fragment | Forward Primer | Reverse Primer |
|---|---|---|---|
| 35S::PDI1: GFP-KDEL | AtPDI1 genomic | ACAACCATGGCTTCGTCATCTACAAGTATCT | CCGACATATGCAACTCATCCTTGGAACTATCA |
| 35S::PDI3: GFP-KDEL | AtPDI3 genomic | ATCAACATGTCGTTAATTCCAAAACCCA | TTACATATGCAATTCATCTTTAGCAGACCCA |
| 35S::PDI4: GFP-KDEL | AtPDI4 genomic | AAACTCGAGTAACAATCATGTTGACGAAACCA | TGGCCCGGGTAACTCATCTTTACCAGACTGATC |
| 35S::PDI5: GFP-KDEL | AtPDI5 genomic | GAACTCGAGAGCGATAATGGCGATGAG | CTCCCCGGGGAGCTCATCCTTGACTTCCT |
| 35S::PDI6: GFP-KDEL | AtPDI6 genomic | AAACTCGAGCGATAATAATGGCGTTTAAG | AGACCCGGGCAGCTCGTCCTTTGCGGCCGT |
| CaMV 35S promoter | TTCAGGGTACCTTCATGGAGTCAAAGATTCA | ATCTACTCGAGTCAAGAGTCCCCCGTG | |
| GFP(S65T)-KDEL:Nos 3'-UTR | CACCGACTAGTCATATGGTGAGCAAGGGCGAG | AGGATGGTCACCTAAAGCTCATCTTTGCCGTGAGTGATC | |
| 35S::PDI9: GFP-KDEL | AtPDI9 genomic | GGGCTCGAGAAAAATGTATAAATCACCATTAAC | GATCCCGGGCAACTCATCCTTAGAACCAAC |
| 35S::PDI10: GFP-KDDL | AtPDI10 genomic | AGGCTCGAGACCATGGAAAGAAAAATGTACAAATCA | AGACATATGCAAGTCGTCCTTGGACTCAGTG |
| GFP(S65T)-KDDL | CACCGACTAGTCATATGGTGAGCAAGGGCGAG | AGGATGGTCACCTAAAGaTCATCTTTGCCGT | |
| 35S::PDI11: GFP-KDEL | AtPDI11 genomic | GAAGCAGAAAAAATCATGACGAAATCTCAG | CAAGCTATATGACATATGAGAAGAAGCAAC |
| 35S::SESA1: mCherry | AtSESA1 genomic | CACACTAGTCAAAAAATGGCAAACAAGTTG | TGAGGATCCGTAGAAAGAAGGGAATGAAG |
| pFLAG-PDI1 | AtPDI1 cDNA | CTTCCCGGGAGAGAATGCGTCCAGTGGATC | CGAAGATCTCAACTCATCCTTGGAACTATCA |
| pFLAG-PDI2 | AtPDI2 cDNA | TTTCCCGGGCGCTTCTTCCTCCGACGAC | TTCGTCGACCAATTCGTCCTTCGAGTCACT |
| pFLAG-PDI3 | AtPDI3 cDNA | CATCCCGGGTTACTCATCACCCGATTCCA | TTAGTCGACCAATTCATCTTTAGCAGACCCA |
| pFLAG-PDI4 | AtPDI4 cDNA | TGTCCCGGGCTCTGATGTCGCCGTCGAAG | TGGGTCGACTAACTCATCTTTACCAGACTGATC |
| pFLAG-PDI5 | AtPDI5 cDNA | TATCCCGGGCGAAGAGACGGAGACGAAG | CTCAGATCTGAGCTCATCCTTGACTTCCTC |
| pFLAG-PDI6 | AtPDI6 cDNA | TATCCCGGGCGAGGAGACGAAGGAATTTG | AGAGTCGACCAGCTCGTCCTTTGCGGCCGT |
| pFLAG-PDI9 | AtPDI9 cDNA | CAGCCCGGGTCTTTATGGATCTTCGTCACCTG | GATgtcgacCAACTCATCCTTAGAACCAACAG |
| pFLAG-PDI10 | AtPDI10 cDNA | GGTGGTACCCTCTATGGATCTTCGTCGCCTG | AGAGTCGACCAAGTCGTCCTTGGACTCAG |
| pFLAG-PDI11 | AtPDI11 cDNA | AGACCCGGGTGACGATGTGGTTGTTTTGACTG | TGAGTCGACAGAAGAAGCAACGAACGTGGTTAG |
| pFLAG-PDI1M | pFLAG-PDI1(a:CGHC) | CTCCGTGGTGCGGCCACTGTCAGGCTTTGAC | GTCAAAGCCTGACAGTGGCCGCACCACGGAG |
| pFLAG-PDI2M | pFLAG-PDI2(a:CGAC) | CTCCGTGGTGTGGTGCTTGTCAGTCTCTTGC | GCAAGAGACTGACAAGCACCACACCACGGAG |
| pFLAG-PDI4M1 | pFLAG-PDI4(a:CARC) | GCATTAGCTCAGCGCACCTCGCACACCAC | GTGGTGTGCGAGGTGCGCTGAGCTAATGC |
| pFLAG-PDI4M2 | pFLAG-PDI4(a:CGHC) | GTTACGCGCCGTGGTGTGGTCATTGCGCTGAGCTAATGCCGAG | CTCGGCATTAGCTCAGCGCAATGACCACACCACGGCGCGTAAC |
| pFLAG-PDI4M3 | pFLAG-PDI4(a′:CGHC) | CACACACCATGGTGTGGTCATTGTGAGGCTCTGAG | CTCAGAGCCTCACAATGACCACACCATGGTGTGTG |
3.2. Protoplast Transient Expression Assays
3.3. Alkaline Phosphatase Activity Assays
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Gething, M.J.; Sambrook, J. Protein folding in the cell. Nature 1992, 355, 33–45. [Google Scholar] [CrossRef]
- Ellis, R.J. The general concept of molecular chaperones. Philos. Trans. R Soc. Lond. B Biol. Sci. 1993, 339, 257–261. [Google Scholar] [CrossRef]
- Miernyk, J.A. Protein folding in the plant cell. Plant Physiol. 1999, 121, 695–703. [Google Scholar] [CrossRef]
- Schröder, M.; Kaufman, R.J. ER stress and the unfolded protein response. Mutat. Res. 2005, 569, 29–63. [Google Scholar] [CrossRef]
- Hebert, D.N.; Molinari, M. In and out of the ER: Protein folding, quality control, degradation, and related human disease. Physiol. Rev. 2007, 87, 1377–1408. [Google Scholar] [CrossRef]
- Freedman, R.B.; Hirst, T.R.; Tuite, M.F. Protein disulphide isomerase: Building bridges in protein folding. Trends Biochem. Sci. 1994, 19, 331–336. [Google Scholar] [CrossRef]
- Edman, J.C.; Ellis, L.; Blacher, R.W.; Roth, R.A.; Rutter, W.J. Sequence of protein disulphide isomerase and implications of its relationship to thioredoxin. Nature 1985, 317, 267–270. [Google Scholar] [CrossRef]
- Kemmink, J.; Darby, N.J.; Dijkstra, K.; Nilges, M.; Creighton, T.E. The folding catalyst protein disulfide isomerase is constructed of active and inactive thioredoxin modules. Curr. Biol. 1997, 7, 239–245. [Google Scholar]
- Wang, C.C.; Tsou, C.L. Protein disulfide isomerase is both an enzyme and a chaperone. FASEB J. 1993, 7, 1515–1517. [Google Scholar]
- Cai, H.; Wang, C.C.; Tsou, C.L. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J. Biol. Chem. 1994, 269, 24550–24552. [Google Scholar]
- Song, J.L.; Wang, C.C. Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. Eur. J. Biochem. 1995, 231, 312–316. [Google Scholar] [CrossRef]
- Hatahet, F.; Ruddock, L.W. Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 2009, 11, 2807–2850. [Google Scholar] [CrossRef]
- Ferrari, D.M.; Söling, H.D. The protein disulphide-isomerase family: Unravelling a string of folds. Biochem. J. 1999, 339, 1–10. [Google Scholar] [CrossRef]
- Kramer, B.; Ferrari, D.M.; Klappa, P.; Pöhlmann, N.; Söling, H.D. Functional roles and efficiencies of the thioredoxin boxes of calcium-binding proteins 1 and 2 in protein folding. Biochem. J. 2001, 357, 83–95. [Google Scholar] [CrossRef]
- Kikuchi, M.; Doi, E.; Tsujimoto, I.; Horibe, T.; Tsujimoto, Y. Functional analysis of human P5, a protein disulfide isomerase homologue. J. Biochem. 2002, 132, 451–455. [Google Scholar] [CrossRef]
- Lu, D.-P.; Christopher, D.A. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genomics 2008, 280, 199–210. [Google Scholar] [CrossRef]
- Li, C.P.; Larkins, B.A. Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol. Biol. 1996, 30, 873–882. [Google Scholar] [CrossRef]
- Takemoto, Y.; Coughlan, S.J.; Okita, T.W.; Satoh, H.; Ogawa, M.; Kumamaru, T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002, 128, 1212–1222. [Google Scholar] [CrossRef]
- Onda, Y.; Nagamine, A.; Sakurai, M.; Kumamaru, T.; Ogawa, M.; Kawagoe, Y. Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell 2011, 23, 210–223. [Google Scholar] [CrossRef]
- Gruber, C.W.; Cemazar, M.; Clark, R.J.; Horibe, T.; Renda, R.F.; Anderson, M.A.; Craik, D.J. A novel plant protein-disulfide isomerase involved in the oxidative folding of cystine knot defense proteins. J. Biol. Chem. 2007, 282, 20435–20446. [Google Scholar]
- Wang, H.; Boavida, L.C.; Ron, M.; McCormick, S. Truncation of a protein disulfide isomerase, PDIL2–1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thalian. Plant Cell 2008, 20, 3300–3311. [Google Scholar] [CrossRef]
- Andème-Ondzighi, C.; Christopher, D.A.; Cho, E.J.; Chang, S.C.; Staehelin, L.A. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 2008, 20, 2205–2220. [Google Scholar] [CrossRef]
- Cho, E.J.; Yuen, C.Y.; Kang, B.H.; Ondzighi, C.A.; Staehelin, L.A.; Christopher, D.A. Protein disulfide isomerase-2 of Arabidopsis mediates protein folding and localizes to both the secretory pathway and nucleus, where it interacts with maternal effect embryo arrest factor. Mol. Cells 2011, 32, 459–475. [Google Scholar] [CrossRef]
- Lu, D.-P.; Christopher, D.A. Immunolocalization of a protein disulfide isomerase to Arabidopsis thaliana chloroplasts and its association with starch biogenesis. Int. J. Plant Sci. 2006, 167, 1–9. [Google Scholar] [CrossRef]
- Kim, J.; Mayfield, S.P. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 1997, 278, 1954–1957. [Google Scholar] [CrossRef]
- Houston, N.L.; Fan, C.; Xiang, J.Q.; Schulze, J.M.; Jung, R.; Boston, R.S. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005, 137, 762–778. [Google Scholar] [CrossRef]
- d’Aloisio, E.; Paolacci, A.R.; Dhanapal, A.P.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. The Protein Disulfide Isomerase gene family in bread wheat (T. aestivum L.). BMC Plant Biol. 2010, 10, e101. [Google Scholar] [CrossRef] [Green Version]
- Lucero, H.A.; Kaminer, B. The role of calcium on the activity of ERcalcistorin/protein-disulfide isomerase and the significance of the C-terminal and its calcium binding. A comparison with mammalian protein-disulfide isomerase. J. Biol. Chem. 1999, 274, 3243–3251. [Google Scholar] [CrossRef]
- Lappi, A.K.; Lensink, M.F.; Alanen, H.I.; Salo, K.E.; Lobell, M.; Juffer, A.H.; Ruddock, L.W. A conserved arginine plays a role in the catalytic cycle of the protein disulphide isomerases. J. Mol. Biol. 2004, 335, 283–295. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Nebenführ, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef]
- Miao, Y.; Li, K.Y.; Li, H.Y.; Yao, X.; Jiang, L. The vacuolar transport of aleurain-GFP and 2S albumin-GFP fusions is mediated by the same pre-vacuolar compartments in tobacco BY-2 and Arabidopsis suspension cultured cells. Plant J. 2008, 56, 824–839. [Google Scholar] [CrossRef]
- Sone, M.; Kishigami, S.; Yoshihisa, T.; Ito, K. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 1997, 272, 6174–6178. [Google Scholar] [CrossRef]
- Humphreys, D.P.; Weir, N.; Mountain, A.; Lund, P.A. Human protein disulfide isomerase functionally complements a dsbA mutation and enhances the yield of pectate lyase C in Escherichia coli. J. Biol. Chem. 1995, 270, 28210–28215. [Google Scholar] [CrossRef]
- Walker, K.W.; Lyles, M.M.; Gilbert, H.F. Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry 1996, 35, 1972–1980. [Google Scholar] [CrossRef]
- Darby, N.J.; Penka, E.; Vincentelli, R. The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J. Mol. Biol. 1998, 276, 239–247. [Google Scholar] [CrossRef]
- Atkin, J.D.; Farg, M.A.; Turner, B.J.; Tomas, D.; Lysaght, J.A.; Nunan, J.; Rembach, A.; Nagley, P.; Beart, P.M.; Cheema, S.S.; Horne, M.K. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J. Biol. Chem. 2006, 281, 30152–30165. [Google Scholar] [CrossRef]
- Solomon, M.; Belenghi, B.; Delledonne, M.; Menachem, E.; Levine, A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 1999, 11, 431–444. [Google Scholar]
- Turano, C.; Coppari, S.; Altieri, F.; Ferraro, A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J. Cell. Physiol. 2002, 193, 154–163. [Google Scholar] [CrossRef]
- Kim, Y.J.; Yeu, S.Y.; Park, B.S.; Koh, H.-J.; Song, J.T.; Seo, H.S. Protein disulfide isomerase-like protein 1-1 controls endosperm development through regulation of the amount and composition of seed proteins in rice. PLoS One 2012, 7, e44493. [Google Scholar]
- Huber-Wunderlich, M.; Glockshuber, R. A single dipeptide sequence modulates the redox properties of a whole enzyme family. Fold. Des. 1998, 3, 161–171. [Google Scholar] [CrossRef]
- Guddat, L.W.; Bardwell, J.C.; Glockshuber, R.; Huber-Wunderlich, M.; Zander, T.; Martin, J.L. Structural analysis of three His32 mutants of DsbA: Support for an electrostatic role of His32 in DsbA stability. Protein Sci. 1997, 6, 1893–1900. [Google Scholar] [CrossRef]
- Jessop, C.E.; Watkins, R.H.; Simmons, J.J.; Tasab, M.; Bulleid, N.J. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J. Cell Sci. 2009, 122, 4287–4295. [Google Scholar] [CrossRef]
- Klappa, P.; Ruddock, L.W.; Darby, N.J.; Freedman, R.B. The b' domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998, 17, 927–935. [Google Scholar] [CrossRef]
- Pagnussat, G.C.; Yu, H.J.; Ngo, Q.A.; Rajani, S.; Mayalagu, S.; Johnson, C.S.; Capron, A.; Xie, L.F.; Ye, D.; Sundaresan, V. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 2005, 132, 603–614. [Google Scholar] [CrossRef]
- Saito, Y.; Kishida, K.; Takata, K.; Takahashi, H.; Shimada, T.; Tanaka, K.; Morita, S.; Satoh, S.; Masumura, T.J. A green fluorescent protein fused to rice prolamin forms protein body-like structures in transgenic rice. Exp. Bot. 2009, 60, 615–627. [Google Scholar] [CrossRef]
- Carrió, M.M.; Villaverde, A. Role of molecular chaperones in inclusion body formation. FEBS Lett. 2003, 537, 215–221. [Google Scholar] [CrossRef]
- Laible, M.; Boonrod, K. Homemade site directed mutagenesis of whole plasmids. J. Vis. Exp. 2009, 11, e1135. [Google Scholar]
- He, P.; Shan, L.; Sheen, J. The use of protoplasts to study innate immune responses. Methods Mol. Biol. 2007, 354, 1–9. [Google Scholar]
- Martínez-García, J.F.; Monte, E.; Quail, P.H. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 1999, 20, 251–257. [Google Scholar] [CrossRef]
- Rietsch, A.; Belin, D.; Martin, N.; Beckwith, J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 1996, 93, 13048–13053. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yuen, C.Y.L.; Matsumoto, K.O.; Christopher, D.A. Variation in the Subcellular Localization and Protein Folding Activity among Arabidopsis thaliana Homologs of Protein Disulfide Isomerase. Biomolecules 2013, 3, 848-869. https://doi.org/10.3390/biom3040848
Yuen CYL, Matsumoto KO, Christopher DA. Variation in the Subcellular Localization and Protein Folding Activity among Arabidopsis thaliana Homologs of Protein Disulfide Isomerase. Biomolecules. 2013; 3(4):848-869. https://doi.org/10.3390/biom3040848
Chicago/Turabian StyleYuen, Christen Y. L., Kristie O. Matsumoto, and David A. Christopher. 2013. "Variation in the Subcellular Localization and Protein Folding Activity among Arabidopsis thaliana Homologs of Protein Disulfide Isomerase" Biomolecules 3, no. 4: 848-869. https://doi.org/10.3390/biom3040848




