Transient Non-Native Helix Formation during the Folding of β-Lactoglobulin
Abstract
:1. Introduction
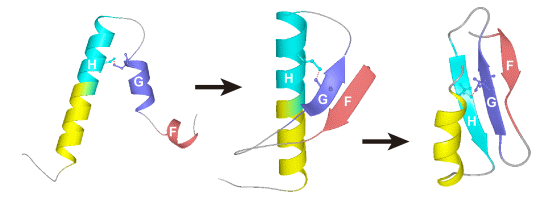
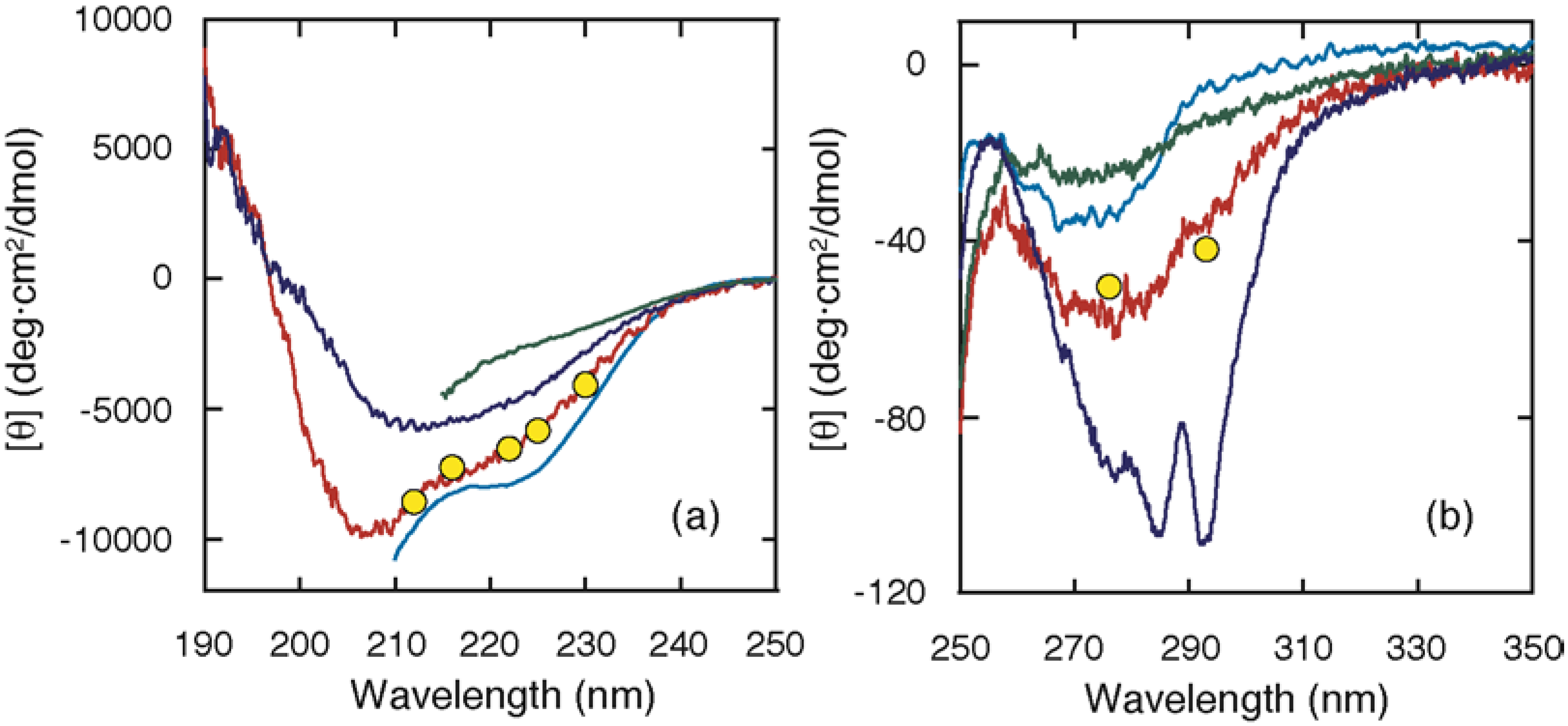
2. Equilibrium Intermediates
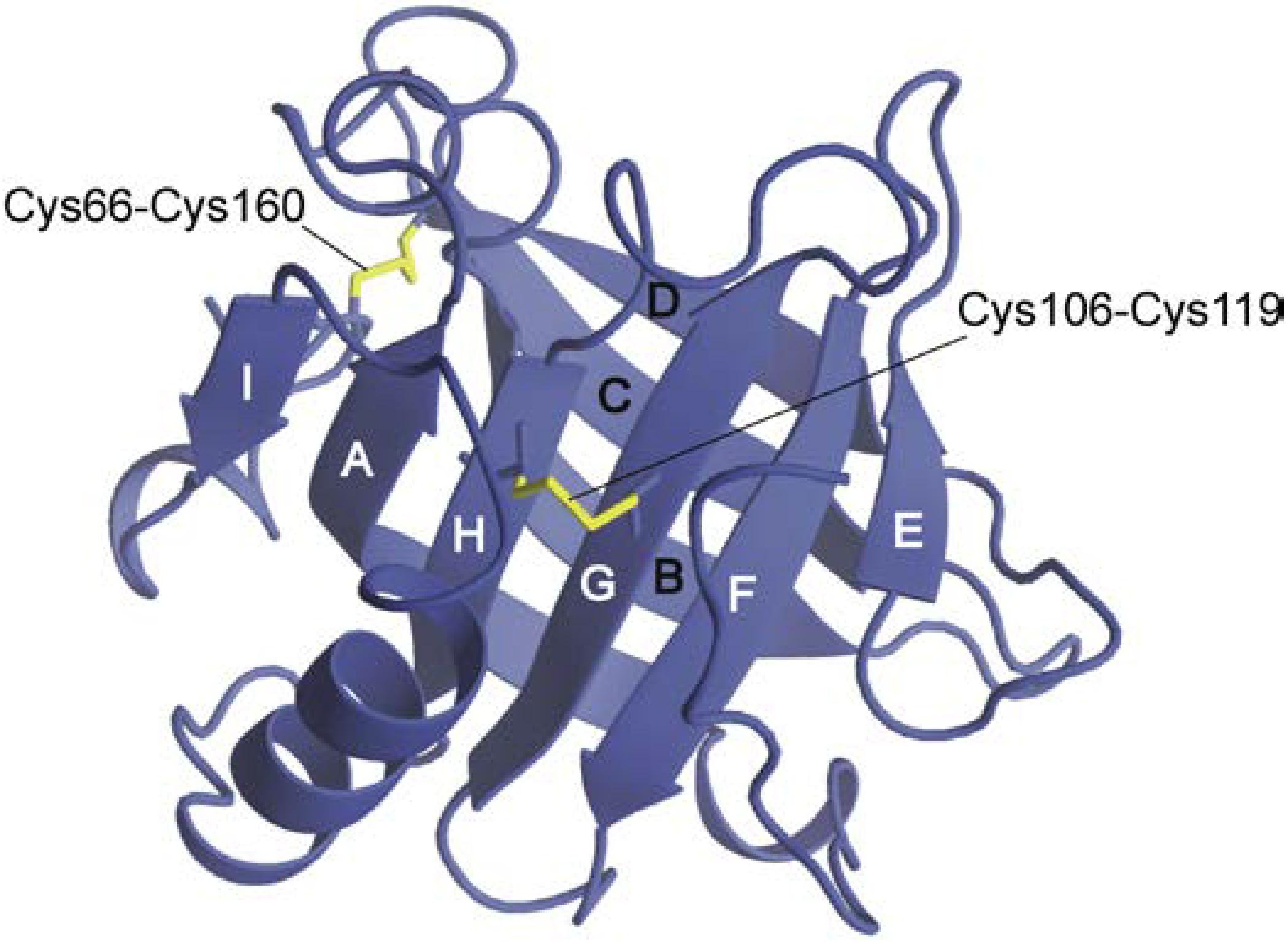
3. Location of Non-Native Helices
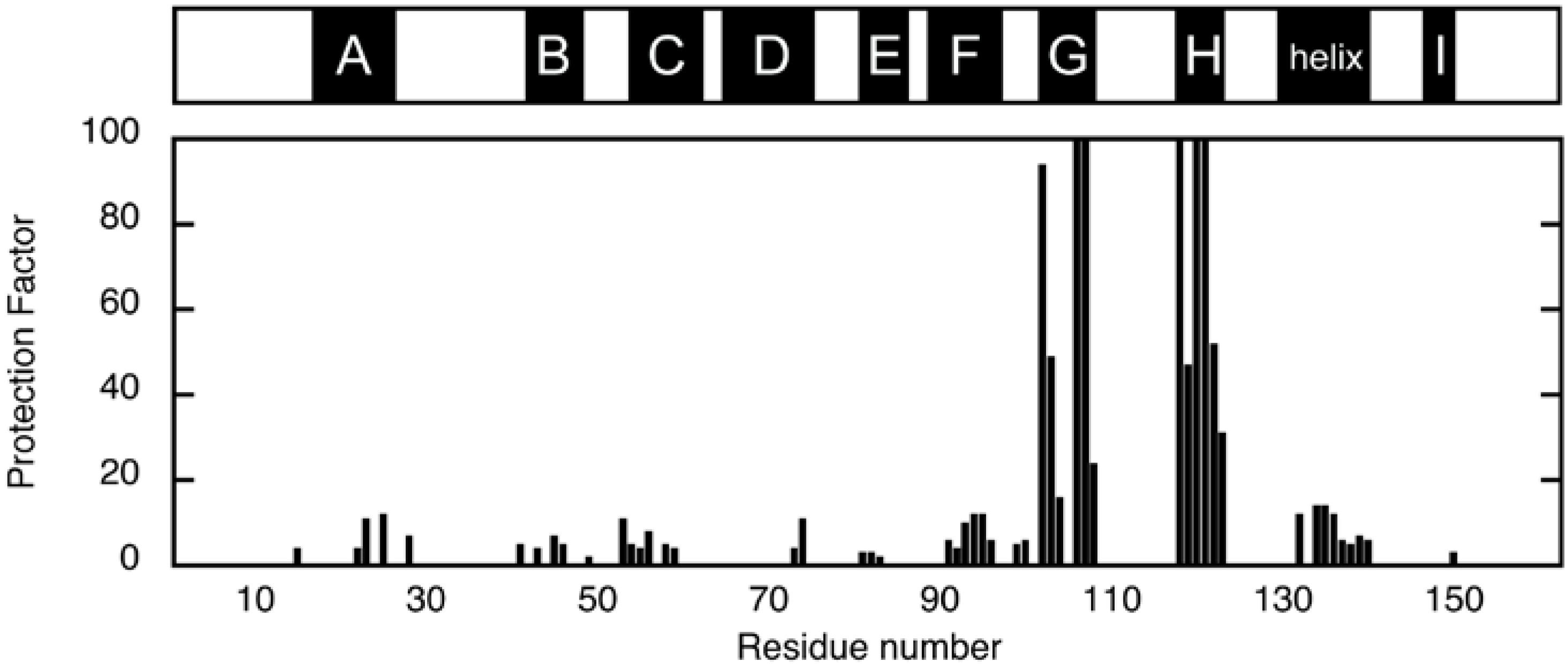
3.1. H/D Exchange
3.2. Proline-Scanning Mutagenesis
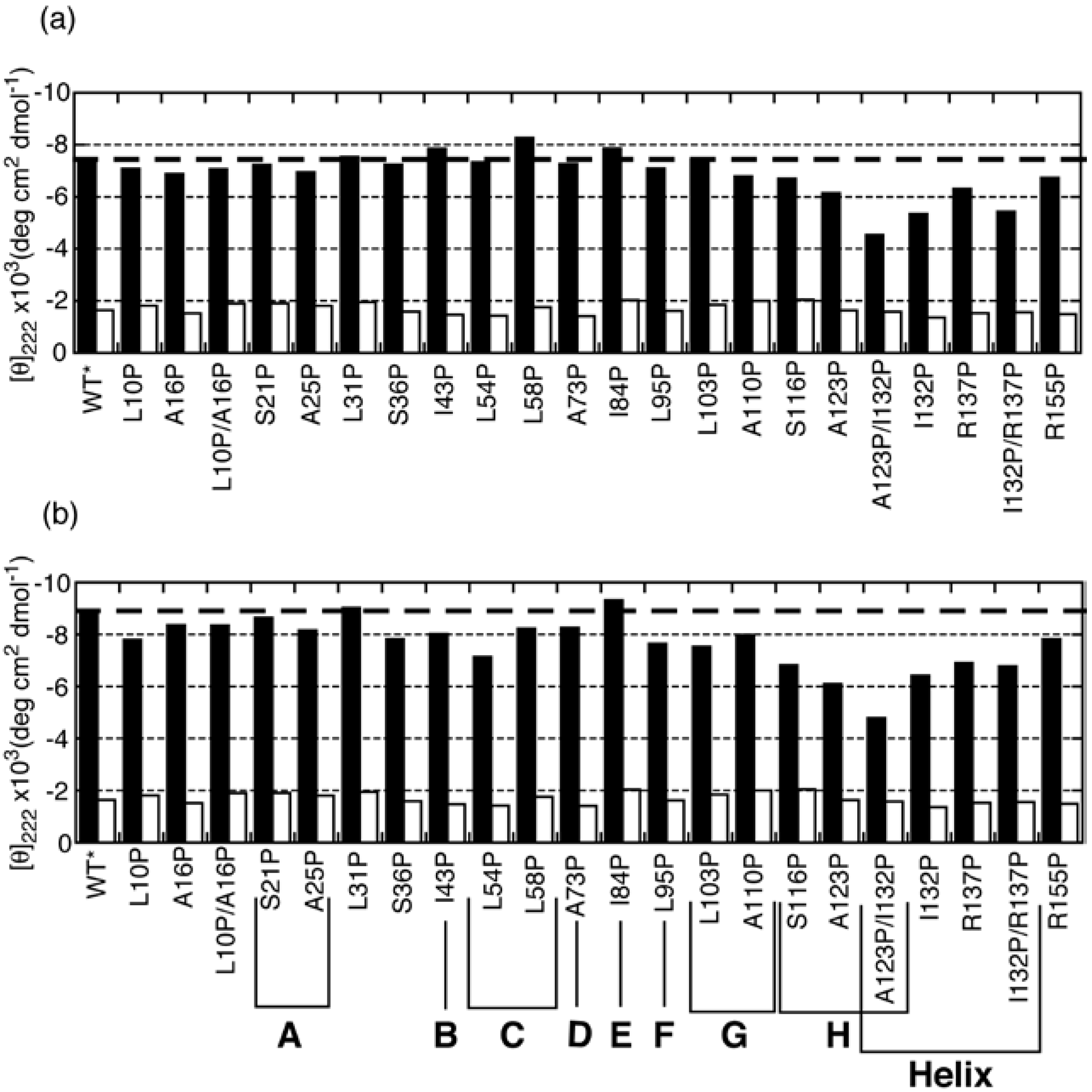
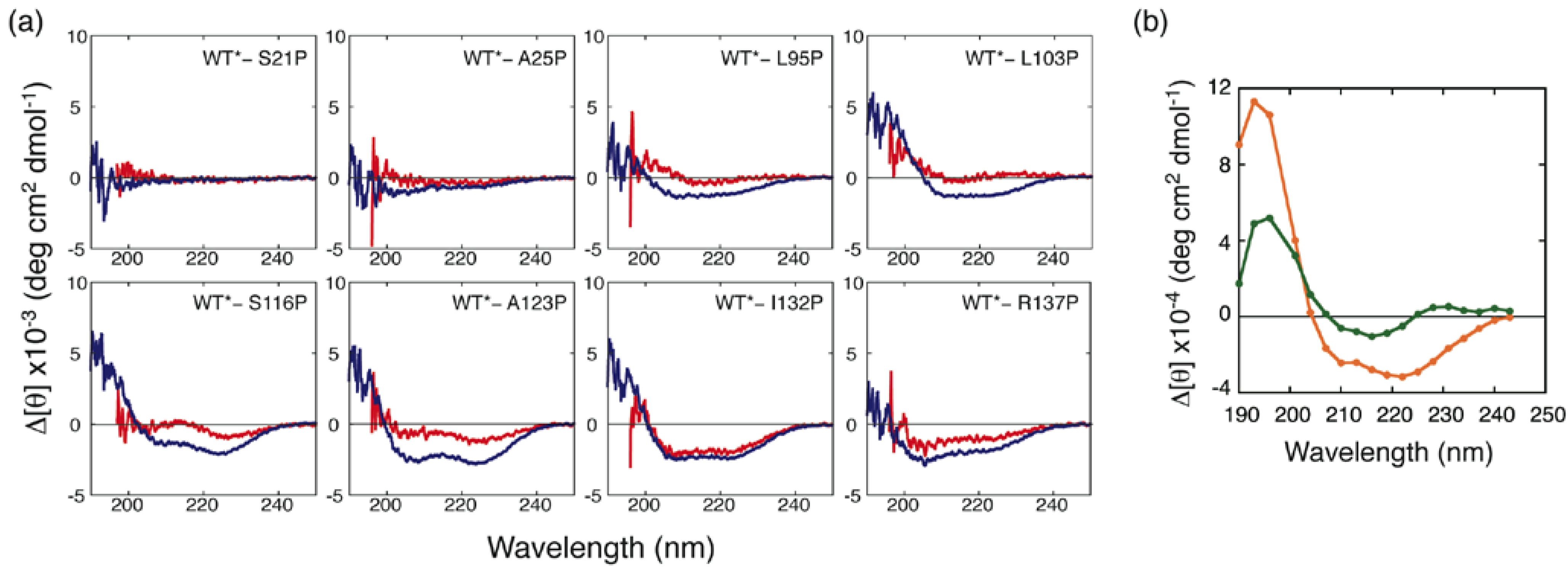
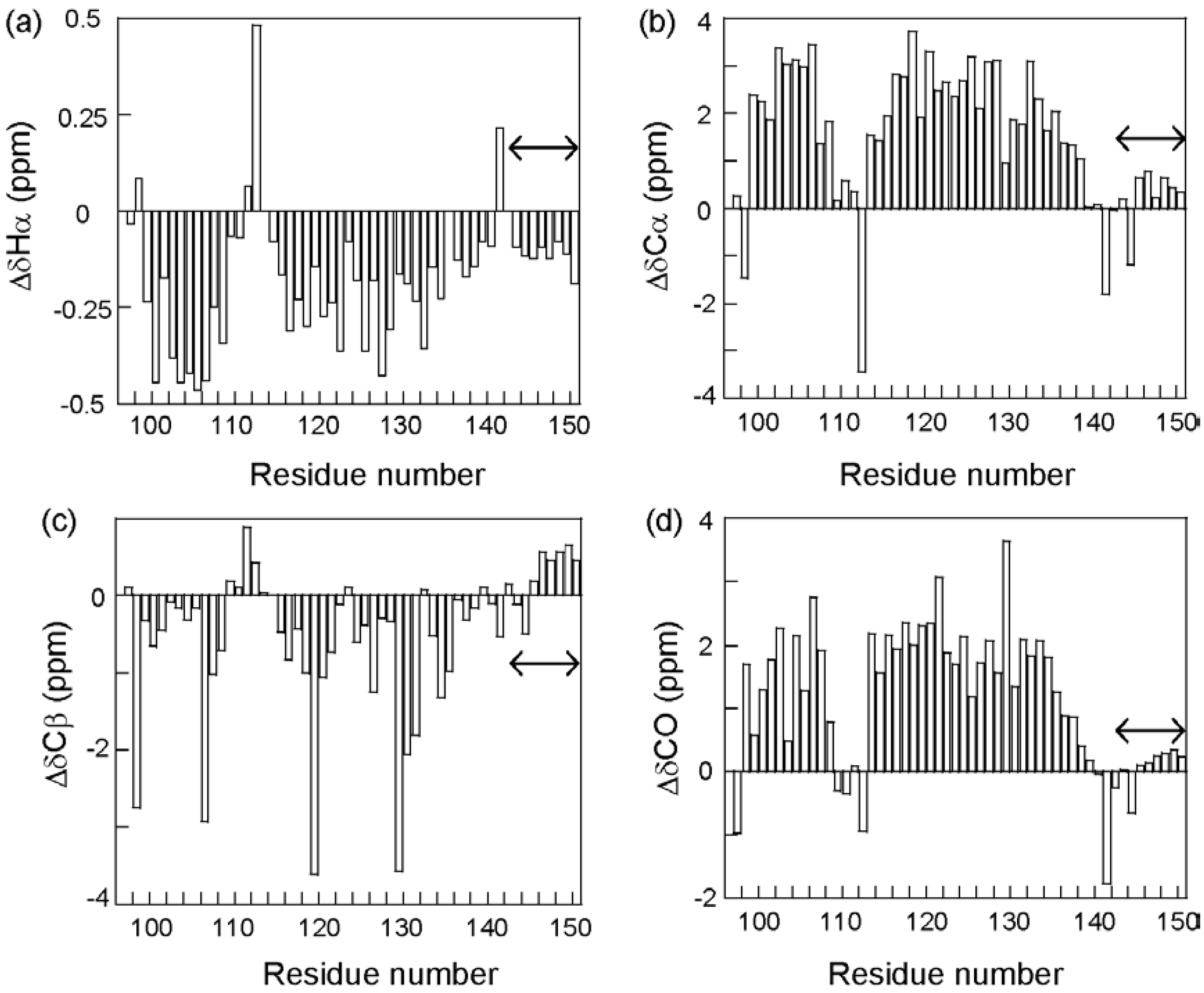
3.3. NMR Analysis of a Fragment
4. Stabilization Mechanisms of Native and Non-Native Structures
4.1. Local Interactions
| Sequence | Corresponding residues | Number of Residues | [θ]MRE f (deg·cm2/dmol) | [θ]M × 10–4 g (deg·cm2/dmol) | Reference |
|---|---|---|---|---|---|
| BLG | 11–18 | 18 | –6400 | [12] | |
| (14–28)GGG(42–52) a | 29 | –3700 | [32] | ||
| 61–77 | 17 | –5500 | [12] | ||
| 85–101 | 16 | –3300 | [12] | ||
| 77–126 | 50 | –9000/–10500 | [32] | ||
| 100–126 | 27 | –3100/–6900 | [32] | ||
| 127–142 | 16 | –6200 | [12] | ||
| ELG | 1–87 | 107 d | –4800 | –51.4 | [33] |
| 88–142 (CHIBL) SS b | 75 d | –10600 | –79.3 | [27] | |
| 88–142 (CHIBL) SH c | 75 d | –6700 | –50.2 | unpublished | |
| 97–142 (CHIBLΔF) SS b | 54 e | –11900 | –64.3 | [28] | |
| 97–142 (CHIBLΔF) SH c | 54 e | –6200 | –33.6 | [28] | |
| 97–110 | 14 | –5500 | –7.7 | [28] | |
| 111–128 | 18 | –4500 | –8.0 | [27] | |
| 111–138 | 28 | –7700 | –21.5 | [28] | |
| 124–138 (F136Y) | 15 | –7200 | –10.8 | [27] |
4.2. A–C Transition
5. Role of the Non-Native Helix
Acknowledgments
Conflict of Interest
References
- Go, N. Theoretical studies of protein folding. Annu. Rev. Biophys. Bioeng. 1983, 12, 183–210. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Wolynes, P.G. Spin glasses and the statistical mechanics of protein folding. Proc. Natl. Acad. Sci. USA 1987, 84, 7524–7528. [Google Scholar] [CrossRef]
- Capaldi, A.P.; Kleanthous, C.; Radford, S.E. Im7 folding mechanism: misfolding on a path to the native state. Nat. Struct. Biol. 2002, 9, 209–216. [Google Scholar]
- Krishna, M.M.; Lin, Y.; Englander, S.W. Protein misfolding: optional barriers, misfolded intermediates, and pathway heterogeneity. J. Mol. Biol. 2004, 343, 1095–1109. [Google Scholar] [CrossRef]
- Nishimura, C.; Dyson, H.J.; Wright, P.E. Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J. Mol. Biol. 2006, 355, 139–156. [Google Scholar] [CrossRef]
- Luheshi, L.M.; Crowther, D.C.; Dobson, C.M. Protein misfolding and disease: from the test tube to the organism. Curr. Opin. Chem. Biol. 2008, 12, 25–31. [Google Scholar] [CrossRef]
- Kuwajima, K.; Yamaya, H.; Miwa, S.; Sugai, S.; Nagamura, T. Rapid formation of secondary structure framework in protein folding studied by stopped-flow circular dichroism. FEBS Lett. 1987, 221, 115–118. [Google Scholar] [CrossRef]
- Qin, B.Y.; Bewley, M.C.; Creamer, L.K.; Baker, H.M.; Baker, E.N.; Jameson, G.B. Structural basis of the Tanford transition of bovine beta-lactoglobulin. Biochemistry 1998, 37, 14014–14023. [Google Scholar] [CrossRef]
- Hamada, D.; Segawa, S.; Goto, Y. Non-native alpha-helical intermediate in the refolding of beta-lactoglobulin, a predominantly beta-sheet protein. Nat. Struct. Biol. 1996, 3, 868–873. [Google Scholar]
- Shiraki, K.; Nishikawa, K.; Goto, Y. Trifluoroethanol-induced stabilization of the alpha-helical structure of beta-lactoglobulin: implication for non-hierarchical protein folding. J. Mol. Biol. 1995, 245, 180–194. [Google Scholar] [CrossRef]
- Hamada, D.; Kuroda, Y.; Tanaka, T.; Goto, Y. High helical propensity of the peptide fragments derived from beta-lactoglobulin, a predominantly beta-sheet protein. J. Mol. Biol. 1995, 254, 737–746. [Google Scholar] [CrossRef]
- Kuroda, Y.; Hamada, D.; Tanaka, T.; Goto, Y. High helicity of peptide fragments corresponding to beta-strand regions of beta-lactoglobulin observed by 2D-NMR spectroscopy. Fold Des. 1996, 1, 255–263. [Google Scholar] [CrossRef]
- Kuwajima, K.; Yamaya, H.; Sugai, S. The burst-phase intermediate in the refolding of beta-lactoglobulin studied by stopped-flow circular dichroism and absorption spectroscopy. J. Mol. Biol. 1996, 264, 806–822. [Google Scholar] [CrossRef]
- Arai, M.; Ikura, T.; Semisotnov, G.V.; Kihara, H.; Amemiya, Y.; Kuwajima, K. Kinetic refolding of beta-lactoglobulin. Studies by synchrotron X-ray scattering, and circular dichroism, absorption and fluorescence spectroscopy. J. Mol. Biol. 1998, 275, 149–162. [Google Scholar] [CrossRef]
- Fujiwara, K.; Arai, M.; Shimizu, A.; Ikeguchi, M.; Kuwajima, K.; Sugai, S. Folding-unfolding equilibrium and kinetics of equine beta-lactoglobulin: equivalence between the equilibrium molten globule state and a burst-phase folding intermediate. Biochemistry 1999, 38, 4455–4463. [Google Scholar] [CrossRef]
- Hamada, D.; Goto, Y. The equilibrium intermediate of beta-lactoglobulin with non-native alpha-helical structure. J. Mol. Biol. 1997, 269, 479–487. [Google Scholar] [CrossRef]
- Kuwata, K.; Hoshino, M.; Era, S.; Batt, C.A.; Goto, Y. alpha-->beta transition of beta-lactoglobulin as evidenced by heteronuclear NMR. J. Mol. Biol. 1998, 283, 731–739. [Google Scholar] [CrossRef]
- Kuwata, K.; Li, H.; Yamada, H.; Batt, C.A.; Goto, Y.; Akasaka, K. High pressure NMR reveals a variety of fluctuating conformers in beta-lactoglobulin. J. Mol. Biol. 2001, 305, 1073–1083. [Google Scholar] [CrossRef]
- Katou, H.; Hoshino, M.; Kamikubo, H.; Batt, C.A.; Goto, Y. Native-like beta-hairpin retained in the cold-denatured state of bovine beta-lactoglobulin. J. Mol. Biol. 2001, 310, 471–484. [Google Scholar] [CrossRef]
- Ikeguchi, M.; Kato, S.; Shimizu, A.; Sugai, S. Molten globule state of equine beta-lactoglobulin. Proteins 1997, 27, 567–575. [Google Scholar] [CrossRef]
- Yamada, Y.; Yajima, T.; Fujiwara, K.; Arai, M.; Ito, K.; Shimizu, A.; Kihara, H.; Kuwajima, K.; Amemiya, Y.; Ikeguchi, M. Helical and expanded conformation of equine beta-lactoglobulin in the cold-denatured state. J. Mol. Biol. 2005, 350, 338–348. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ikeguchi, M.; Sugai, S. Molten globule structure of equine beta-lactoglobulin probed by hydrogen exchange. J. Mol. Biol. 2000, 299, 757–770. [Google Scholar] [CrossRef]
- Kuwata, K.; Shastry, R.; Cheng, H.; Hoshino, M.; Batt, C.A.; Goto, Y.; Roder, H. Structural and kinetic characterization of early folding events in beta-lactoglobulin. Nat. Struct. Biol. 2001, 8, 151–155. [Google Scholar]
- Nakagawa, K.; Tokushima, A.; Fujiwara, K.; Ikeguchi, M. Proline scanning mutagenesis reveals non-native fold in the molten globule state of equine beta-lactoglobulin. Biochemistry 2006, 45, 15468–15473. [Google Scholar] [CrossRef]
- Yamada, Y.; Nakagawa, K.; Yajima, T.; Saito, K.; Tokushima, A.; Fujiwara, K.; Ikeguchi, M. Structural and thermodynamic consequences of removal of a conserved disulfide bond from equine beta-lactoglobulin. Proteins 2006, 63, 595–602. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yang, J.T.; Chau, K.H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 1974, 13, 3350–3359. [Google Scholar] [CrossRef]
- Nakagawa, K.; Yamada, Y.; Fujiwara, K.; Ikeguchi, M. Interactions responsible for secondary structure formation during folding of equine beta-lactoglobulin. J. Mol. Biol. 2007, 367, 1205–1214. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nakagawa, K.; Fujiwara, K.; Shimizu, A.; Ikeguchi, M. A native disulfide stabilizes non-native helical structures in partially folded states of equine beta-lactoglobulin. Biochemistry 2011, 50, 10590–10597. [Google Scholar]
- Wishart, D.S. Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 58, 62–87. [Google Scholar] [CrossRef]
- Zimm, B.H.; Bragg, J.K. Theory of the Phase Transition between Helix and Random Coil in Polypeptide Chains. J. Chem. Phys. 1959, 31, 526–535. [Google Scholar] [CrossRef]
- Lifson, S.; Roig, A. On the Theory of Helix---Coil Transition in Polypeptides. J Chem Phys 1961, 34, 1963–1974. [Google Scholar] [CrossRef]
- Ragona, L.; Catalano, M.; Zetta, L.; Longhi, R.; Fogolari, F.; Molinari, H. Peptide models of folding initiation sites of bovine beta-lactoglobulin: identification of nativelike hydrophobic interactions involving G and H strands. Biochemistry 2002, 41, 2786–2796. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nakagawa, K.; Ikeguchi, M. Importance of polypeptide chain length for the correct local folding of a beta-sheet protein. Biophys. Chem. 2012, 168–169, 40–47. [Google Scholar] [CrossRef]
- Nakagawa, K.; Yamada, Y.; Matsumura, Y.; Tsukamoto, S.; Yamamoto-Ohtomo, M.; Ohtomo, H.; Okabe, T.; Fujiwara, K.; Ikeguchi, M. Relationship between chain collapse and secondary structure formation in a partially folded protein. Biopolymers 2014, in press. [Google Scholar]
- Wensley, B.G.; Batey, S.; Bone, F.A.; Chan, Z.M.; Tumelty, N.R.; Steward, A.; Kwa, L.G.; Borgia, A.; Clarke, J. Experimental evidence for a frustrated energy landscape in a three-helix-bundle protein family. Nature 2010, 463, 685–688. [Google Scholar] [CrossRef]
- Prigozhin, M.B.; Liu, Y.; Wirth, A.J.; Kapoor, S.; Winter, R.; Schulten, K.; Gruebele, M. Misplaced helix slows down ultrafast pressure-jump protein folding. Proc. Natl. Acad. Sci. USA 2013, 110, 8087–8092. [Google Scholar]
- Chikenji, G.; Kikuchi, M. What is the role of non-native intermediates of beta-lactoglobulin in protein folding? Proc. Natl. Acad. Sci. USA 2000, 97, 14273–14277. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Yamashita, T.; Yamada, Y.; Fujiwara, K.; Maki, K.; Kuwajima, K.; Matsumura, Y.; Kihara, H.; Tsuge, H.; Ikeguchi, M. Non-native alpha-helix formation is not necessary for folding of lipocalin: Comparison of burst-phase folding between tear lipocalin and beta-lactoglobulin. Proteins 2009, 76, 226–236. [Google Scholar] [CrossRef]
- Sakurai, K.; Fujioka, S.; Konuma, T.; Yagi, M.; Goto, Y. A Circumventing Role for the Non-Native Intermediate in the Folding of beta-Lactoglobulin. Biochemistry 2011, 50, 6498–6507. [Google Scholar] [CrossRef]
- Qin, Z.; Hu, D.; Shimada, L.; Nakagawa, T.; Arai, M.; Zhou, J.M.; Kihara, H. Refolding of beta-lactoglobulin studied by stopped-flow circular dichroism at subzero temperatures. FEBS Lett. 2001, 507, 299–302. [Google Scholar] [CrossRef]
- Larios, E.; Li, J.S.; Schulten, K.; Kihara, H.; Gruebele, M. Multiple probes reveal a native-like intermediate during low-temperature refolding of ubiquitin. J. Mol. Biol. 2004, 340, 115–125. [Google Scholar] [CrossRef]
- Li, J.S.; Shinjo, M.; Matsumura, Y.; Morita, M.; Baker, D.; Ikeguchi, M.; Kihara, H. An alpha-helical burst in the src SH3 folding pathway. Biochemistry 2007, 46, 5072–5082. [Google Scholar] [CrossRef]
- Matsumura, Y.; Shinjo, M.; Kim, S.J.; Okishio, N.; Gruebele, M.; Kihara, H. Transient helical structure during PI3K and Fyn SH3 domain folding. J. Phys. Chem. B 2013, 117, 4836–4843. [Google Scholar]
- Matsumura, Y.; Shinjo, M.; Mahajan, A.; Tsai, M.D.; Kihara, H. alpha-Helical burst on the folding pathway of FHA domains from Rad53 and Ki67. Biochimie 2010, 92, 1031–1039. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ikeguchi, M. Transient Non-Native Helix Formation during the Folding of β-Lactoglobulin. Biomolecules 2014, 4, 202-216. https://doi.org/10.3390/biom4010202
Ikeguchi M. Transient Non-Native Helix Formation during the Folding of β-Lactoglobulin. Biomolecules. 2014; 4(1):202-216. https://doi.org/10.3390/biom4010202
Chicago/Turabian StyleIkeguchi, Masamichi. 2014. "Transient Non-Native Helix Formation during the Folding of β-Lactoglobulin" Biomolecules 4, no. 1: 202-216. https://doi.org/10.3390/biom4010202