Structure and Function of the Bi-Directional Bacterial Flagellar Motor
Abstract
:1. Introduction
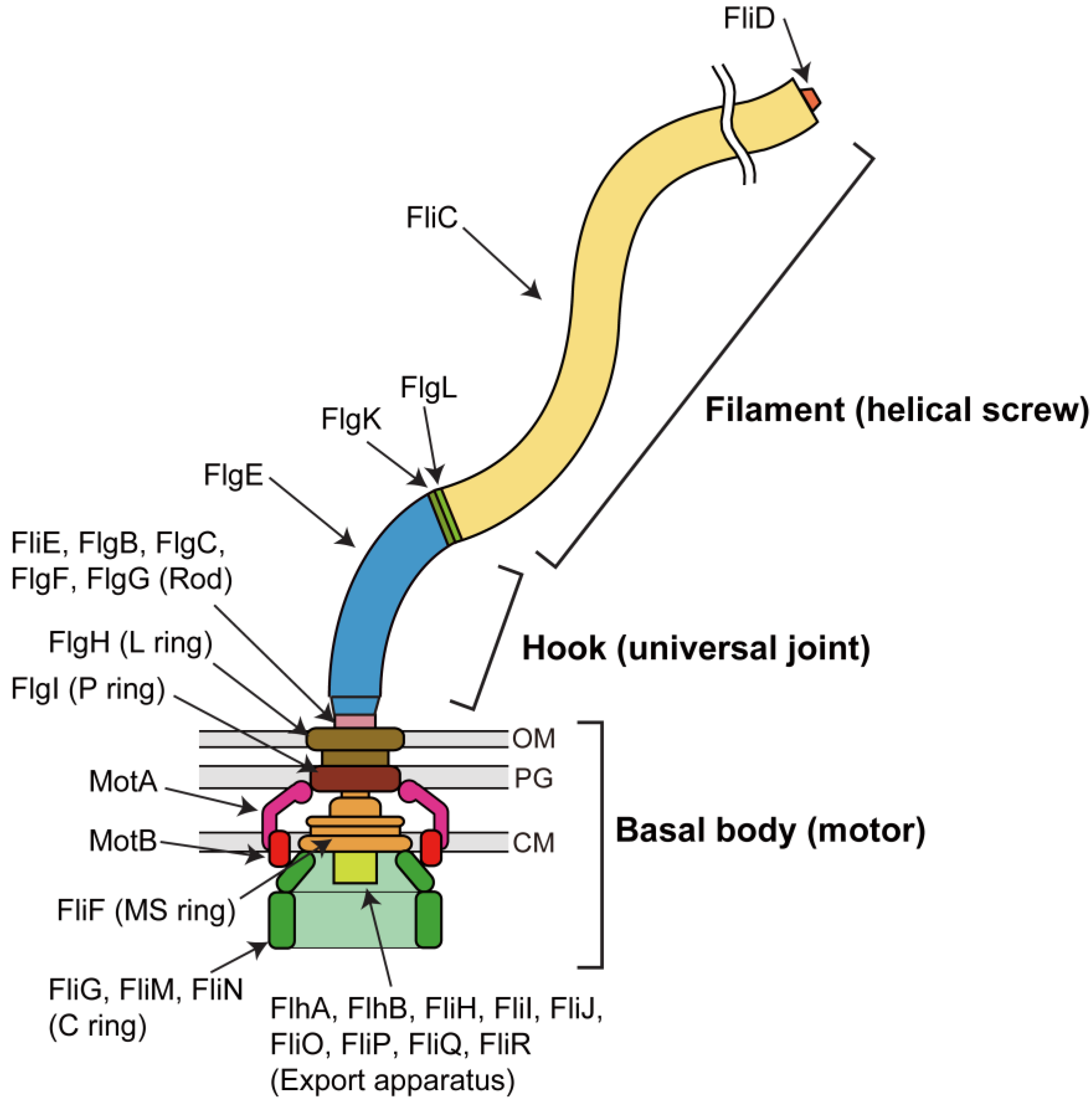
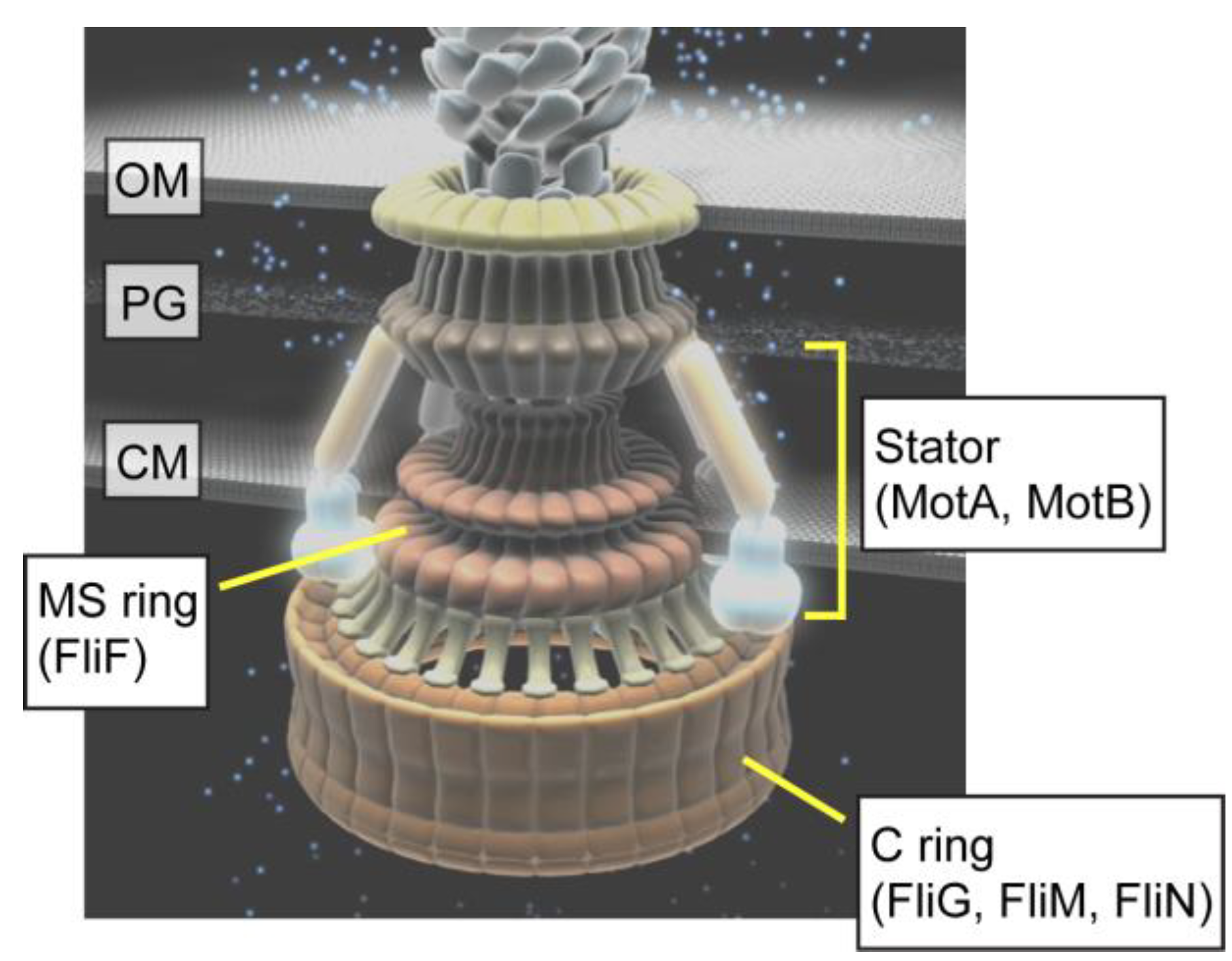
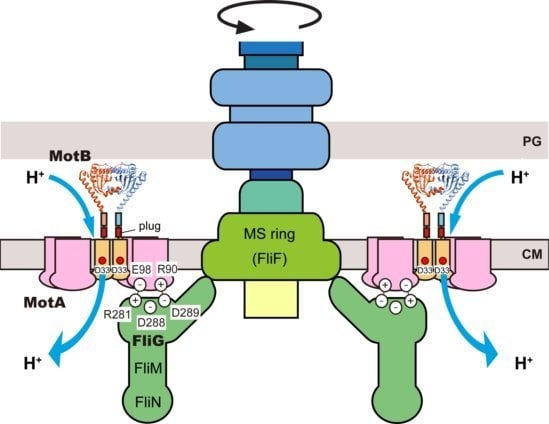
2. Rotor
3. Stator
3.1. Proton Channel of the MotA/B Complex

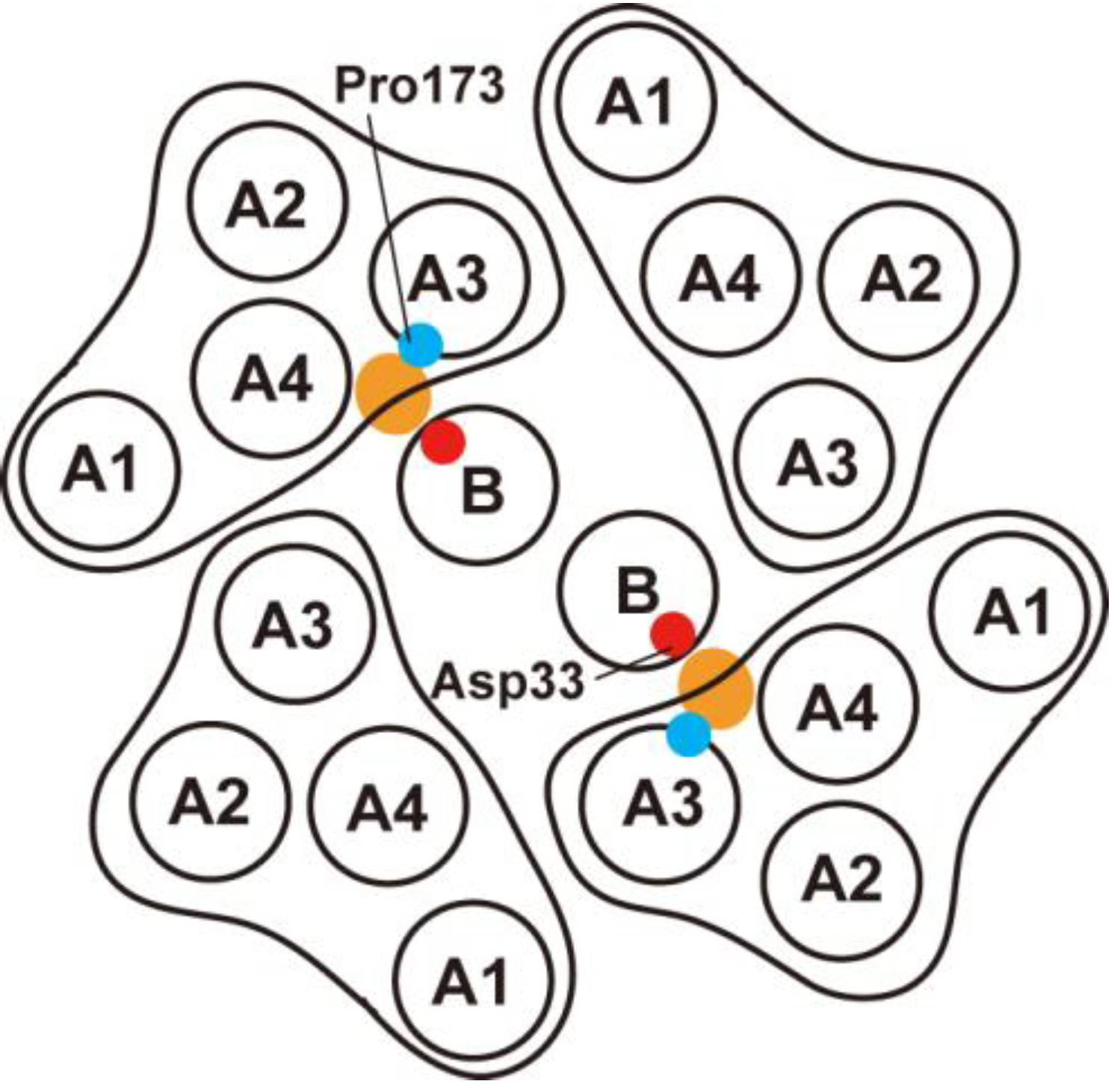
3.2. Role of MotBC in Stator Assembly around a Rotor
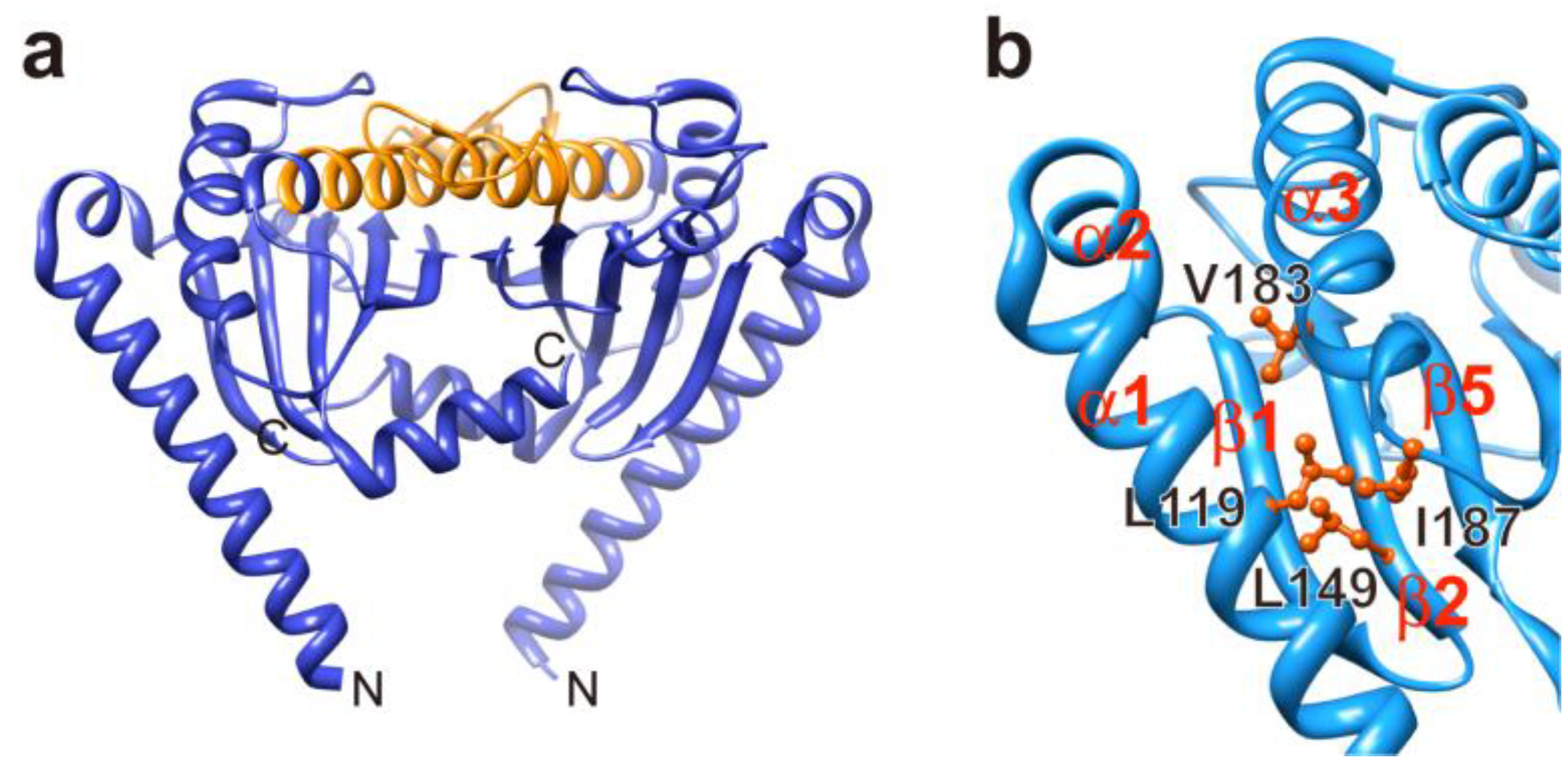
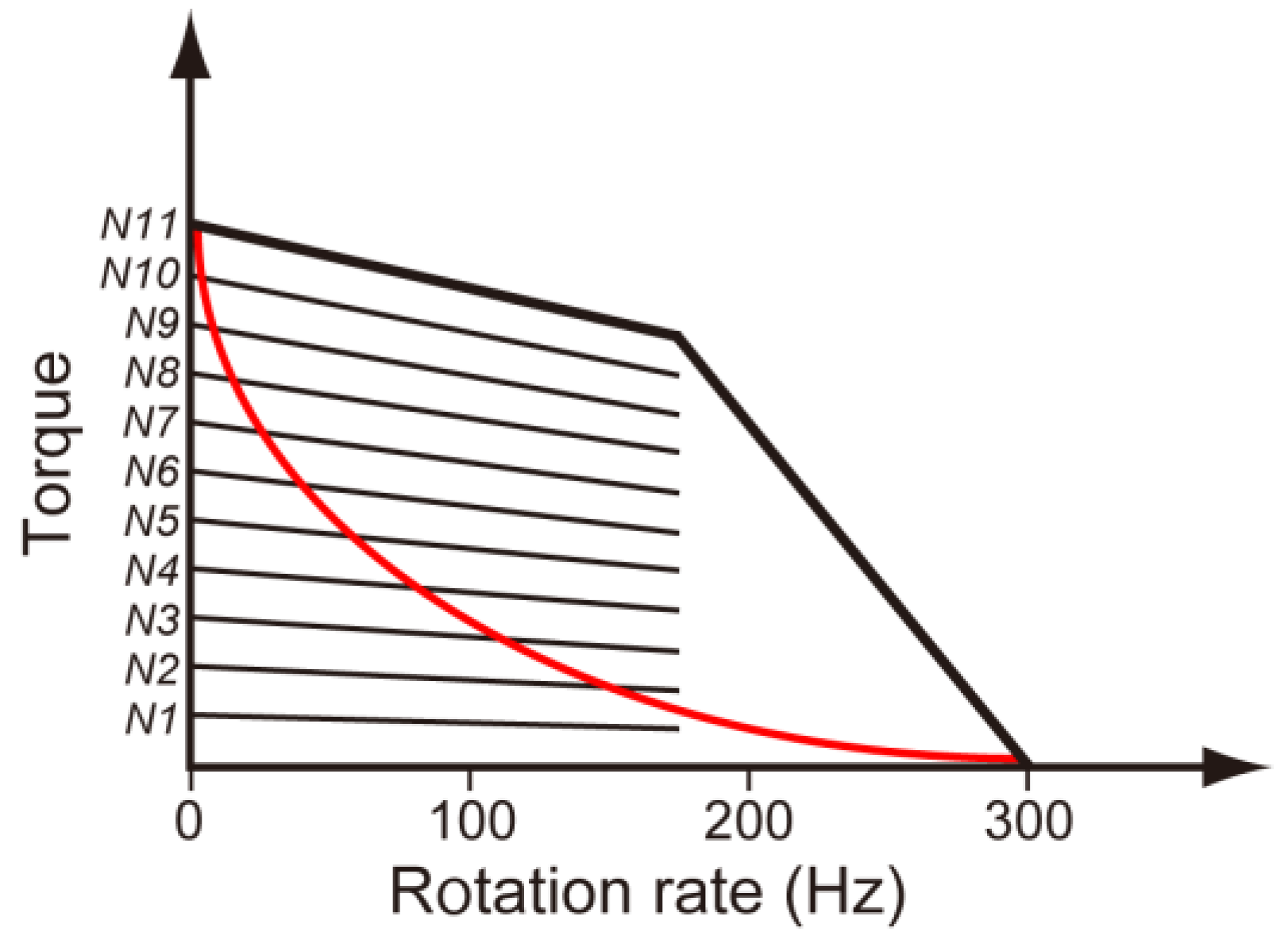
3.3. Load-Sensitive Coupling between Proton Translocation and Torque Generation
4. Role of Stator-Rotor Interactions in Efficient Assembly of the Stators into the Motor
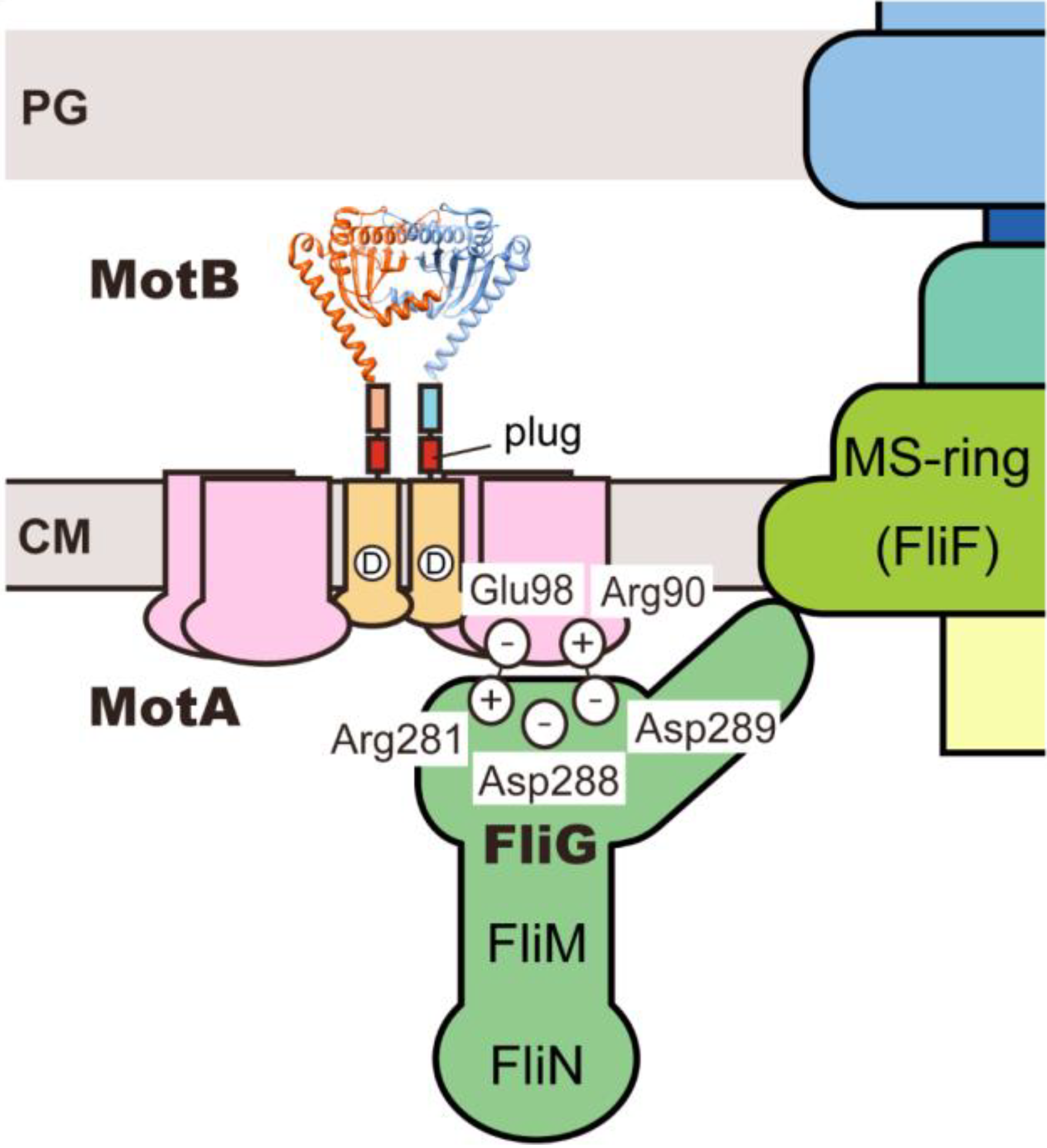
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Berg, H.C. The rotary motor of bacterial flagella. Ann. Rev. Biochem. 2003, 72, 19–54. [Google Scholar] [CrossRef]
- Macnab, R.M. How bacteria assemble flagella. Annu. Rev. Microbiol. 2003, 57, 77–100. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K.; Namba, K. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 2008, 18, 693–701. [Google Scholar] [CrossRef]
- Blair, D.F. Flagellar movement driven by proton translocation. FEBS Lett. 2003, 545, 86–95. [Google Scholar] [CrossRef]
- Sowa, Y.; Berry, R.M. Bacterial flagellar motor. Q. Rev. Biophys. 2008, 41, 103–132. [Google Scholar]
- Macnab, R.M.; Ornston, M.K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J. Mol. Biol. 1977, 112, 1–30. [Google Scholar] [CrossRef]
- Turner, L.; Ryu, W.S.; Berg, H.C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 2000, 182, 2793–2801. [Google Scholar]
- Sourjik, V.; Armitage, J.P. Spatial organization in bacterial chemotaxis. EMBO J. 2010, 29, 2724–2733. [Google Scholar] [CrossRef]
- Ueno, T.; Oosawa, K.; Aizawa, S. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 1992, 227, 672–677. [Google Scholar] [CrossRef]
- Suzuki, H.; Yonekura, K.; Namba, K. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 2004, 337, 105–113. [Google Scholar] [CrossRef]
- Khan, I.H.; Reese, T.S.; Khan, S. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 1992, 89, 5956–5960. [Google Scholar] [CrossRef]
- Francis, N.R.; Sosinsky, G.E.; Thomas, D.; DeRosier, D.J. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 1994, 235, 1261–1270. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Aizawa, S.; Kihara, M.; Isomura, M.; Jones, C.J.; Macnab, R.M. Genetic evidence for a switching and energytransducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 1986, 168, 1172–1179. [Google Scholar]
- Block, S.M.; Berg, H.C. Successive incorporation of force generating units in the bacterial rotary motor. Nature 1984, 309, 470–472. [Google Scholar] [CrossRef]
- Blair, D.F.; Berg, H.C. Restoration of torque in defective flagellar motors. Science 1988, 242, 1678–1681. [Google Scholar]
- Wilson, M.L.; Macnab, R.M. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J. Bacteriol. 1990, 172, 3932–3939. [Google Scholar]
- De Mot, R.; Vanderleyden, J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 1994, 12, 333–334. [Google Scholar] [CrossRef]
- Blair, D.F.; Berg, H.C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 1990, 60, 439–449. [Google Scholar] [CrossRef]
- Stolz, B.; Berg, H.C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 1991, 173, 7033–7037. [Google Scholar]
- Tang, H.; Braun, T.F.; Blair, D.F. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 1996, 261, 209–221. [Google Scholar] [CrossRef]
- Morimoto, Y.V.; Che, Y.-S.; Minamino, T.; Namba, K. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS lett. 2010, 584, 1268–1272. [Google Scholar] [CrossRef]
- Zhou, J.; Lloyd, S.A.; Blair, D.F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 1998, 95, 6436–6441. [Google Scholar] [CrossRef]
- Francis, N.R.; Irikura, V.M.; Yamaguchi, S.; DeRosier, D.J.; Macnab, R.M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 1992, 89, 6304–6308. [Google Scholar] [CrossRef]
- Oosawa, K.; Ueno, T.; Aizawa, S. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 1994, 176, 3683–3691. [Google Scholar]
- Lee, L.K.; Ginsburg, M.A.; Crovace, C.; Donohoe, M.; Stock, D. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 2010, 466, 996–1000. [Google Scholar] [CrossRef]
- Kihara, M.; Miller, G.U.; Macnab, R.M. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 2000, 182, 3022–3028. [Google Scholar] [CrossRef]
- Levenson, R.; Zhou, H.; Dahlquist, F.W. Structural insights into the interaction between the bacterial flagellar motor proteins FliF and FliG. Biochemistry 2012, 51, 5052–5060. [Google Scholar] [CrossRef]
- Brown, P.N.; Terrazas, M.; Paul, K.; Blair, D.F. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 2007, 189, 305–312. [Google Scholar] [CrossRef]
- Paul, K.; Gonzalez-Bonet, G.; Bilwes, A.M.; Crane, B.R.; Blair, D. Architecture of the flagellar rotor. EMBO J. 2011, 30, 2962–2971. [Google Scholar] [CrossRef]
- Vartanian, A.S.; Paz, A.; Fortgang, E.A.; Abramson, J.; Dahlquist, F.W. Structure of flagellar motor proteins in complex allows for insights into motor structure and switching. J. Biol. Chem. 2012, 287, 35779–35783. [Google Scholar]
- Lam, K.H.; Lam, W.W.; Wong, J.Y.; Chan, L.C.; Kotaka, M.; Ling, T.K.; Jin, D.Y.; Ottemann, K.M.; Au, S.W. Structural basis of FliG-FliM interaction in Helicobacter pylori. Mol Microbiol. 2013, 88, 798–812. [Google Scholar] [CrossRef]
- Brown, P.N.; Mathews, M.A.; Joss, L.A.; Hill, C.P.; Blair, D.F. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 2005, 187, 2890–2902. [Google Scholar] [CrossRef]
- Thomas, D.R.; Morgan, D.G.; DeRosier, D.J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 1999, 96, 10134–10139. [Google Scholar] [CrossRef]
- Thomas, D.R.; Francis, N.R.; Xu, C.; DeRosier, D.J. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006, 188, 7039–7048. [Google Scholar] [CrossRef]
- Lloyd, S.A.; Whitby, F.G.; Blair, D.F.; Hill, C.P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 1999, 400, 472–475. [Google Scholar] [CrossRef]
- Brown, P.N.; Hill, C.P.; Blair, D.F. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002, 21, 3225–3234. [Google Scholar] [CrossRef]
- Minamino, T.; Imada, K.; Kinoshita, M.; Nakamura, S.; Morimoto, Y.V.; Namba, K. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 2011, 9, e1000616. [Google Scholar] [CrossRef]
- Park, S.Y.; Lowder, B.; Bilwes, A.M.; Blair, D.F.; Crane, B.R. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. USA 2006, 103, 11886–11891. [Google Scholar]
- Sircar, R.; Greenswag, A.R.; Bilwes, A.M.; Gonzalez-Bonet, G.; Crane, B.R. Structure and activity of the flagellar rotor protein FliY: a member of the CheC phosphatase family. J. Biol. Chem. 2013, 288, 13493–13502. [Google Scholar]
- Stock, D.; Namba, K.; Lee, L.K. Nanorotors and self-assembling macromolecular machines: the torque ring of the bacterial flagellar motor. Curr. Opin. Biotechnol. 2012, 23, 545–554. [Google Scholar] [CrossRef]
- Dyer, C.M.; Vartanian, A.S.; Zhou, H.; Dahlquist, F.W. A molecular mechanism of bacterial flagellar motor switching. J. Mol. Biol. 2009, 388, 71–84. [Google Scholar] [CrossRef]
- Sarkar, M.K.; Paul, K.; Blair, D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2010, 107, 9370–9375. [Google Scholar] [CrossRef]
- Cluzel, P.; Surette, M.; Leibler, S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 2000, 287, 1652–1655. [Google Scholar] [CrossRef]
- Duke, T.A.; Le Novere, N.; Bray, D. Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 2001, 308, 541–553. [Google Scholar] [CrossRef]
- Bai, F.; Branch, R.W.; Nicolau, D.V., Jr.; Pilizota, T.; Steel, B.C.; Maini, P.K.; Berry, R.M. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science 2010, 327, 685–689. [Google Scholar]
- Yuan, J.; Fahrner, K.A.; Berg, H.C. Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 2009, 390, 394–400. [Google Scholar] [CrossRef]
- Bai, F.; Minamino, T.; Wu, Z.; Namba, K.; Xing, J. Coupling between switching regulation and torque generation in bacterial flagellar motor. Phys. Rev. Lett. 2012, 108, 178105. [Google Scholar] [CrossRef]
- Boehm, A.; Kaiser, M.; Li, H.; Spangler, C.; Kasper, C.A.; Ackermann, M.; Kaever, V.; Sourjik, V.; Roth, V.; Jenal, U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 2010, 141, 1–10. [Google Scholar]
- Paul, K.; Nieto, V.; Carlquist, W.C.; Blair, D.F.; Harshey, R.M. (2010) The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a "backstop brake" mechanism. Mol. Cell. 2010, 38, 128–139. [Google Scholar] [CrossRef]
- Fang, X.; Gomelsky, M. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 2010, 76, 1295–1305. [Google Scholar] [CrossRef]
- Van Way, S.M.; Millas, S.G.; Lee, A.H.; Manson, M.D. Rusty, jammed, and well-oiled hinges: Mutations affecting the interdomain region of FliG, a rotor element of the Escherichia coli flagellar motor. J. Bacteriol. 2004, 186, 3173–3181. [Google Scholar] [CrossRef]
- Togashi, F.; Yamaguchi, S.; Kihara, M.; Aizawa, S.-I.; Macnab, R.M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 1997, 179, 2994–3003. [Google Scholar]
- Sowa, Y.; Rowe, A.D.; Leake, M.C.; Yakushi, T.; Homma, M.; Ishijima, A.; Berry, R.M. Direct observation of steps in rotation of the bacterial flagellar motor. Nature 2005, 437, 916–919. [Google Scholar] [CrossRef]
- Nakamura, S.; Kami-ike, N.; Yokota, J.P.; Minamino, T.; Namba, K. Evidence for symmetry in the elementary process of bidirectional torque generation by the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2010, 107, 17616–17620. [Google Scholar]
- Yuan, J.; Fahrner, K.A.; Turner, L.; Berg, H.C. Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2010, 107, 12846–12849. [Google Scholar] [CrossRef]
- Delalez, N.J.; Wadhams, G.H.; Rosser, G.; Xue, Q.; Brown, M.T.; Dobbie, I.M.; Berry, R.M.; Leake, M.C.; Armitage, J.P. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc. Natl. Acad. Sci. USA 2010, 107, 11347–11351. [Google Scholar] [CrossRef]
- Fukuoka, H.; Inoue, Y.; Terasawa, S.; Takahashi, H.; Ishijima, A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem. Biophys. Res. Commun. 2010, 394, 130–135. [Google Scholar]
- Yuan, J.; Branch, R.W.; Hosu, B.G.; Berg, H.C. Adaptation at the output of the chemotaxis signalling pathway. Nature 2012, 484, 233–236. [Google Scholar]
- Yuan, J.; Berg, H.C. Ultrasensitivity of an adaptive bacterial motor. J. Mol. Biol. 2013, 425, 1760–1764. [Google Scholar] [CrossRef]
- Larsen, S.H.; Adler, J.; Gargus, J.J.; Hogg, R.W. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl. Acad. Sci. USA 1974, 71, 1239–1243. [Google Scholar] [CrossRef]
- Manson, M.D.; Tedesco, P.; Berg, H.C.; Harold, F.M.; Van der Drift, C. A protonmotive force drives bacterial flagella. Proc. Natl. Acad. Sci. USA 1977, 4, 3060–3064. [Google Scholar]
- Matsuura, S.; Shioi, J.; Imae, Y. Motility in Bacillus subtilis driven by an artificial protonmotive force. FEBS Lett. 1977, 82, 187–190. [Google Scholar] [CrossRef]
- Ravid, S.; Eisenbach, M. Minimal requirements for rotation of bacterial flagella. J. Bacteriol. 1984, 158, 1208–1210. [Google Scholar]
- Terahara, N.; Sano, M.; Ito, M. A Bacillus flagellar motor that can use both Na+ and K+ as a coupling ion is converted by a single mutation to use only Na+. PLoS One 2012, 7, e46248. [Google Scholar] [CrossRef]
- Sharp, L.L.; Zhou, J.; Blair, D.F. Tryptophan scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry 1995, 34, 9166–9171. [Google Scholar] [CrossRef]
- Zhou, J.; Sharp, L.L.; Tang, H.L.; Lloyd, S.A.; Billings, S.; Braun, T.F.; Blair, D.F. Function of protonatable residues in the flagellar motor of Escherichia coli: A critical role for Asp 32 of MotB. J. Bacteriol. 1998, 180, 2729–2735. [Google Scholar]
- Hirota, N.; Kitada, M.; Imae, Y. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+. FEBS Lett. 1981, 132, 278–280. [Google Scholar] [CrossRef]
- Hirota, N.; Imae, Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain Yn-1. J. Biol. Chem. 1983, 258, 10577–10581. [Google Scholar]
- Chernyak, B.V.; Dibrov, P.A.; Glagolev, A.N.; Sherman, M.Y.; Skulachev, V.P. A novel type of energetics in a marine alkali-tolerant bacterium: Δμ-Na-driven motility and sodium cycle. FEBS Lett. 1983, 164, 38–42. [Google Scholar] [CrossRef]
- Kojima, S.; Blair, D.F. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 2004, 43, 26–34. [Google Scholar] [CrossRef]
- Terahara, N.; Krulwich, T.A.; Ito, M. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl. Acad. Sci. USA 2008, 105, 14359–14364. [Google Scholar] [CrossRef]
- Li, N.; Kojima, S.; Homma, M. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells. 2011, 16, 985–999. [Google Scholar]
- Sato, K.; Homma, M. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 2000, 275, 5718–5722. [Google Scholar] [CrossRef]
- Terashima, H.; Fukuoka, H.; Yakushi, T.; Kojima, S.; Homma, M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol. Microbiol. 2006, 62, 1170–1180. [Google Scholar] [CrossRef]
- Ito, M.; Hicks, D.B.; Henkin, T.M.; Guffanti, A.A.; Powers, B.D.; Zvi, L.; Uematsu, K.; Krulwich, T.A. MotPS is the stator-force generator for motility of alkaliphilic Bacillus and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 2004, 53, 1035–1049. [Google Scholar] [CrossRef]
- Paulick, A.; Koerdt, A.; Lassak, J.; Huntley, S.; Wilms, I.; Narberhaus, F.; Thormann, K.M. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol. Microbiol. 2009, 71, 836–850. [Google Scholar] [CrossRef]
- Hosking, E.R.; Manson, M.D. Clusters of charged residues at the C terminus of MotA and N terminus of MotB are important for function of the Escherichia coli flagellar motor. J. Bacteriol. 2008, 190, 5517–5521. [Google Scholar] [CrossRef]
- Braun, T.; Blair, D.F. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry 2001, 40, 13051–13059. [Google Scholar] [CrossRef]
- Braun, T.F.; Al-Mawasawi, L.Q.; Kojima, S.; Blair, D.F. Arrangement of core membrane segments in the MotA/MotB protein-channel complex of Escherichia coli. Biochemistry 2004, 43, 35–45. [Google Scholar] [CrossRef]
- Che, Y.-S.; Nakamura, S.; Kojima, S.; Kami-ike, N.; Namba, K.; Minamino, T. Suppressor analysis of the MotB(D33E) mutation to probe the bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 2008, 190, 6660–6667. [Google Scholar] [CrossRef]
- Kim, E.A.; Price-Carter, M.; Carlquist, W.C.; Blair, D.F. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry 2008, 47, 11332–11339. [Google Scholar] [CrossRef]
- Blair, D.F.; Berg, H.C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J. Mol. Biol. 1991, 221, 1433–1442. [Google Scholar] [CrossRef]
- Braun, T.; Poulson, S.; Gully, J.B.; Empey, J.C.; Van Way, S.; Putnam, A.; Blair, D.F. Function of proline residues of MotA in torque generation by the flagellar motor of Escherichia coli. J. Bacteriol. 1999, 181, 3542–3551. [Google Scholar]
- Nakamura, S.; Morimoto, Y.V.; Kami-ike, N.; Minamino, T.; Namba, K. Role of a conserved prolyl residue (Pro-173) of MotA in the mechanochemical reaction cycle of the proton-driven flagellar motor of Salmonella. J. Mol. Biol. 2009, 393, 300–307. [Google Scholar] [CrossRef]
- Brandl, C.J.; Deber, C.M. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc. Natl. Acad. Sci. USA 1986, 83, 917–921. [Google Scholar] [CrossRef]
- Hosking, E.R.; Vogt, C.; Bakker, E.P.; Manson, M.D. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 2006, 364, 921–937. [Google Scholar] [CrossRef]
- Muramoto, K.; Macnab, R.M. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 1998, 29, 1191–202. [Google Scholar] [CrossRef]
- Kojima, S.; Imada, K.; Sakuma, M.; Sudo, Y.; Kojima, C.; Minamino, T.; Homma, M.; Namba, K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009, 73, 710–718. [Google Scholar] [CrossRef]
- Kojima, S.; Furukawa, Y.; Matsunami, H.; Minamino, T.; Namba, K. Characterization of the periplasmic domain of MotB and implications for its role in the stator assembly of the bacterial flagellar motor. J. Bacteriol. 2008, 190, 2259–2266. [Google Scholar]
- Blair, D.F.; Kim, D.Y.; Berg, H.C. Mutant MotB proteins in Escherichia coli. J. Bacteriol. 1991, 173, 4049–4055. [Google Scholar]
- Garza, A.G.; Biran, R.; Wohlschlegel, J.; Manson, M.D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J. Mol. Biol. 1996, 25, 270–285. [Google Scholar]
- Hizukuri, Y.; Morton, J.F.; Yakushi, T.; Kojima, S.; Homma, M. The peptidoglycan-binding (PGB) domain of the Escherichia coli pal protein can also function as the PGB domain in E. coli flagellar motor protein MotB. J. Biochem. 2009, 146, 219–229. [Google Scholar] [CrossRef]
- Hizukuri, Y.; Kojima, S.; Homma, M. Disulfide cross-linking between the stator and the bearing components in the bacterial flagellar motor. J. Biochem. 2010, 148, 309–318. [Google Scholar] [CrossRef]
- Ryu, W.S.; Berry, R.M.; Berg, H.C. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 2000, 403, 444–447. [Google Scholar] [CrossRef]
- Yuan, J.; Berg, H.C. Resurrection of the flagellar rotary motor near zero load. Proc. Natl Acad. Sci. USA 2008, 105, 1182–1185. [Google Scholar] [CrossRef]
- Reid, S.W.; Leake, M.C.; Chandler, J.H.; Lo, C.J.; Armitage, J.P.; Berry, R.M. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. USA 2006, 103, 8066–8071. [Google Scholar]
- Leake, M.C.; Chandler, J.H.; Wadhams, G.H.; Bai, F.; Berry, R.M.; Armitage, J.P. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 2006, 443, 355–358. [Google Scholar] [CrossRef]
- Lele, P.P.; Hosu, B.G.; Berg, H.C. Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 2013, 110, 11839–11844. [Google Scholar] [CrossRef]
- Tipping, M.J.; Delalez, N.J.; Lim, R.; Berry, R.M.; Armitage, J.P. Load-dependent assembly of the bacterial flagellar motor. MBio 2013, 4, e00551-13. [Google Scholar]
- Castillo, D.J.; Nakamura, S.; Morimoto, Y.V.; Che, Y.-S.; Kamiike, N.; Kudo, S.; Minamino, T.; Namba, K. The C-terminal periplasmic domain of MotB is responsible for load-dependent control of the number of stators of the bacterial flagellar motor. Biophysics 2013, 9, 173–181. [Google Scholar] [CrossRef]
- Kojima, S.; Blair, D.F. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 2001, 40, 13041–13050. [Google Scholar] [CrossRef]
- Che, Y.-S.; Nakamura, S.; Morimoto, Y.V.; Kami-ike, N.; Namba, K.; Minamino, T. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol. Microbiol. 2014, 91, 175–184. [Google Scholar] [CrossRef]
- Chen, X.; Berg, H.C. Solvent-isotope and pH effects on flagellar rotation in Escherichia coli. Biophys. J. 2000, 78, 2280–2284. [Google Scholar] [CrossRef]
- Zhou, J.; Blair, D.F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 1997, 273, 428–439. [Google Scholar] [CrossRef]
- Morimoto, Y.V.; Nakamura, S.; Kami-ike, N.; Namba, K.; Minamino, T. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 2010, 78, 1117–1129. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, Y.V.; Nakamura, S.; Hiraoka, K.D.; Namba, K.; Minamino, T. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J. Bacteriol. 2013, 195, 474–481. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Morimoto, Y.V.; Minamino, T. Structure and Function of the Bi-Directional Bacterial Flagellar Motor. Biomolecules 2014, 4, 217-234. https://doi.org/10.3390/biom4010217
Morimoto YV, Minamino T. Structure and Function of the Bi-Directional Bacterial Flagellar Motor. Biomolecules. 2014; 4(1):217-234. https://doi.org/10.3390/biom4010217
Chicago/Turabian StyleMorimoto, Yusuke V., and Tohru Minamino. 2014. "Structure and Function of the Bi-Directional Bacterial Flagellar Motor" Biomolecules 4, no. 1: 217-234. https://doi.org/10.3390/biom4010217