Structures and Metal-Binding Properties of Helicobacter pylori Neutrophil-Activating Protein with a Di-Nuclear Ferroxidase Center
Abstract
:1. Introduction
2. Overall Structures of HP-NAP
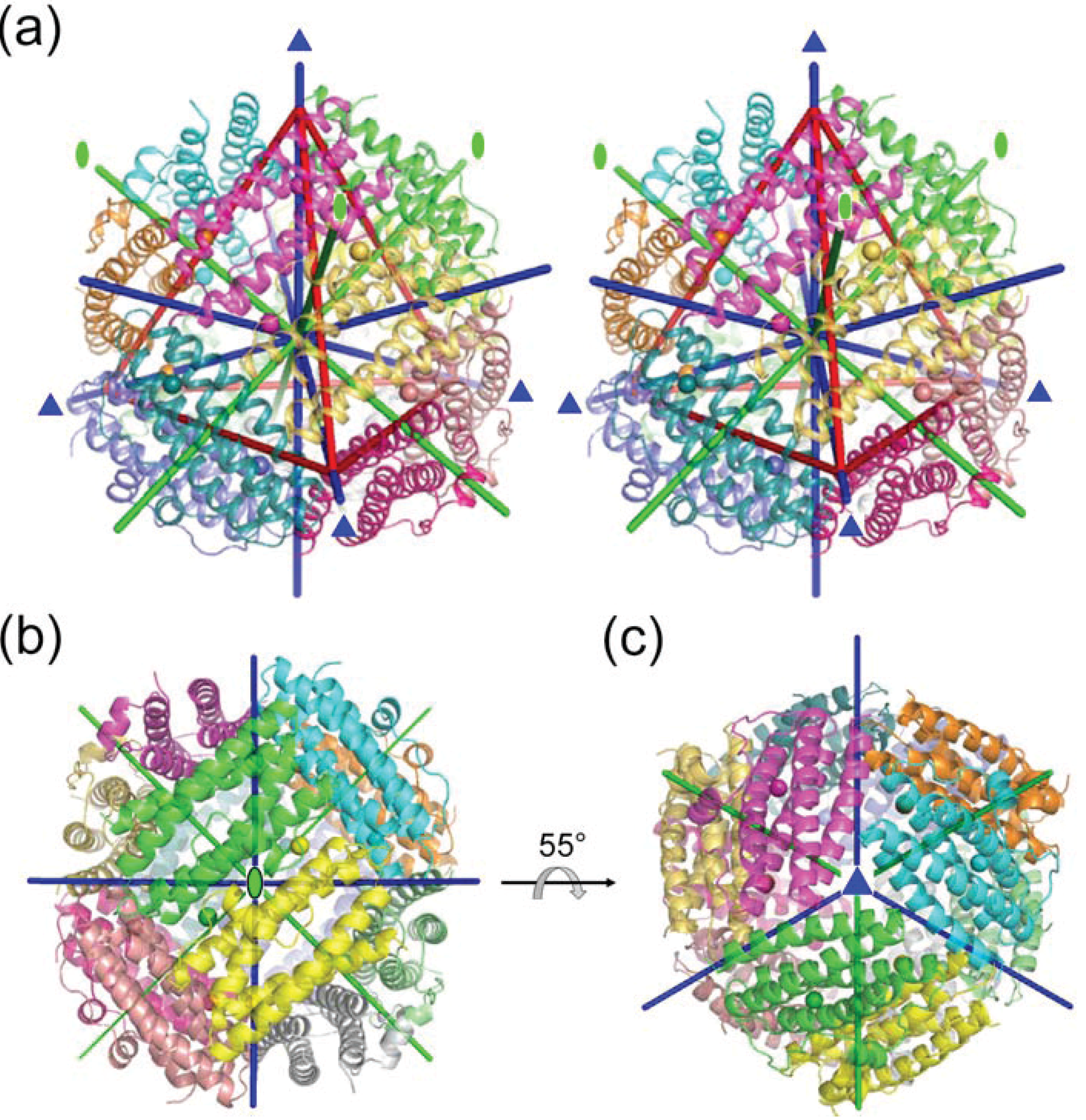
2.1. Dodecameric Structure
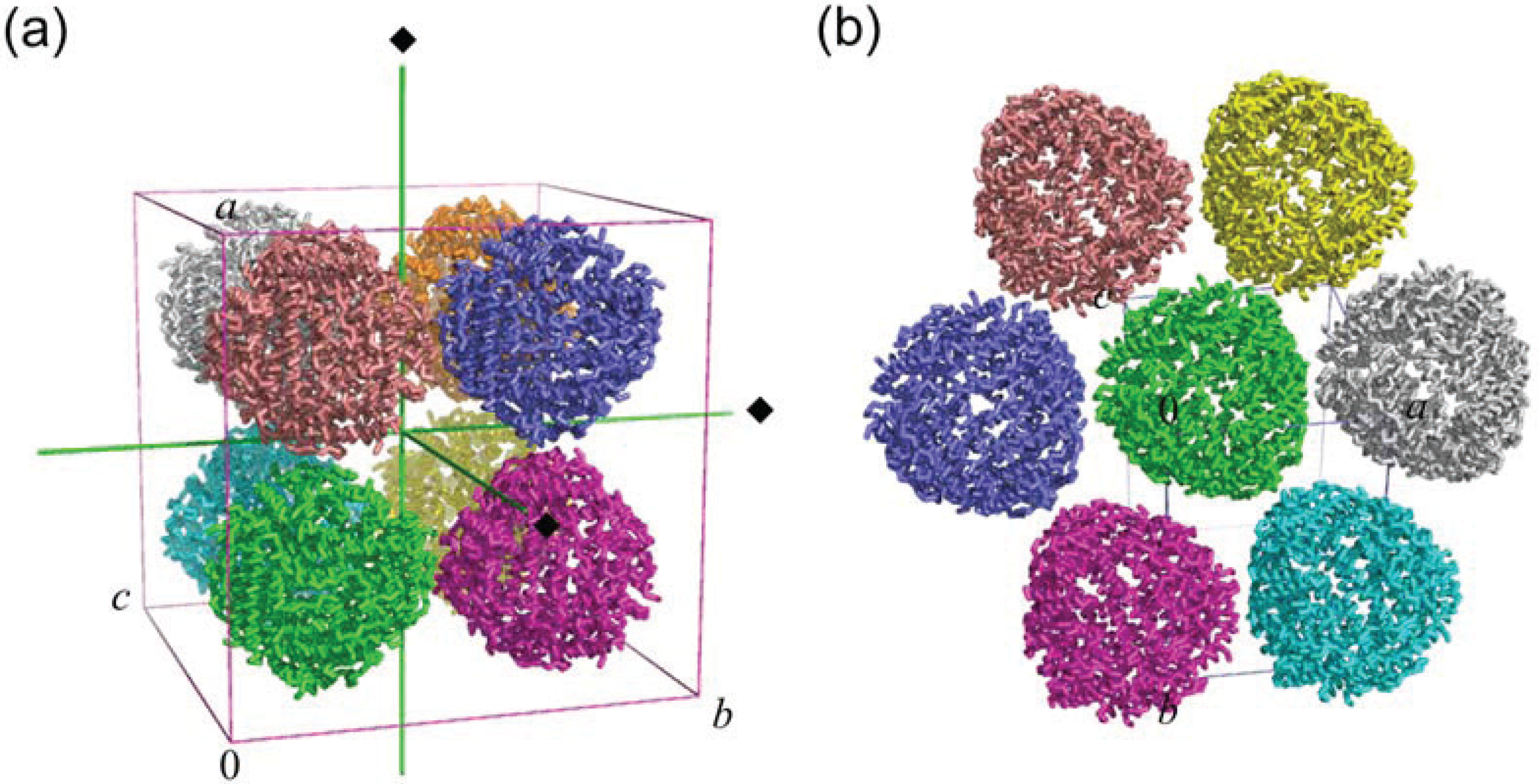
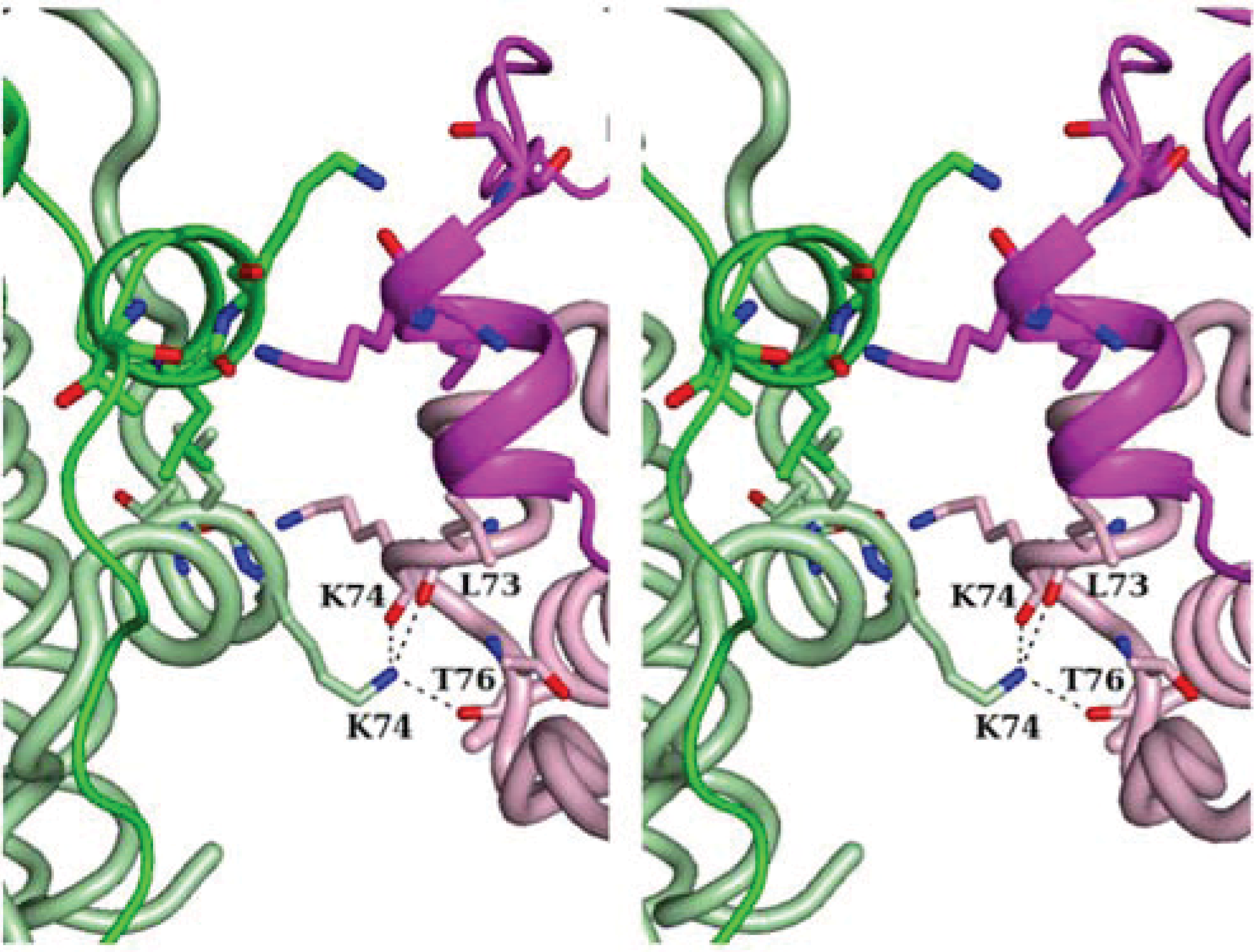
2.2. Crystal Packing
3. Ferroxidase Center
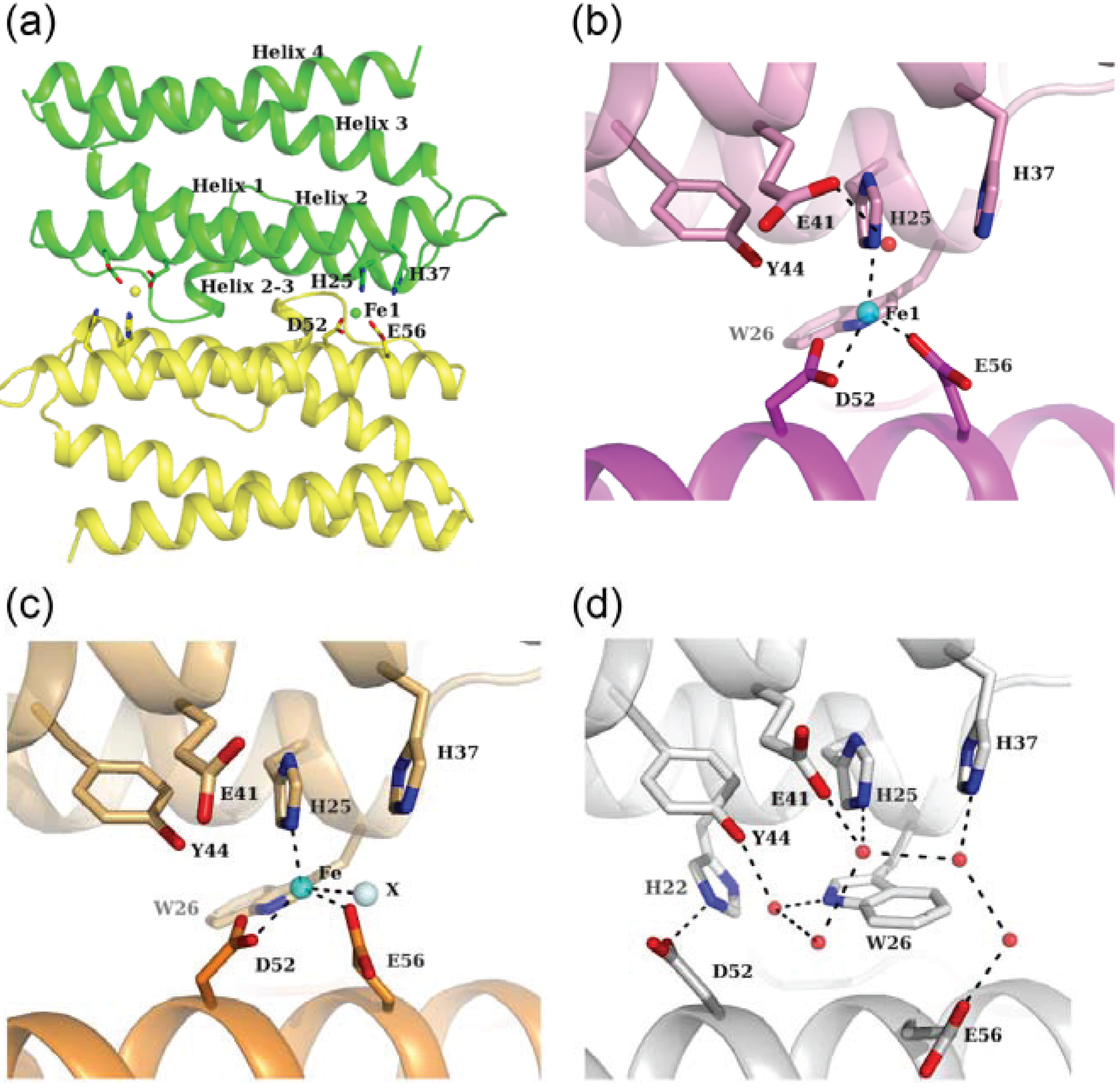
3.1. Fe-Loaded and Apo Structures
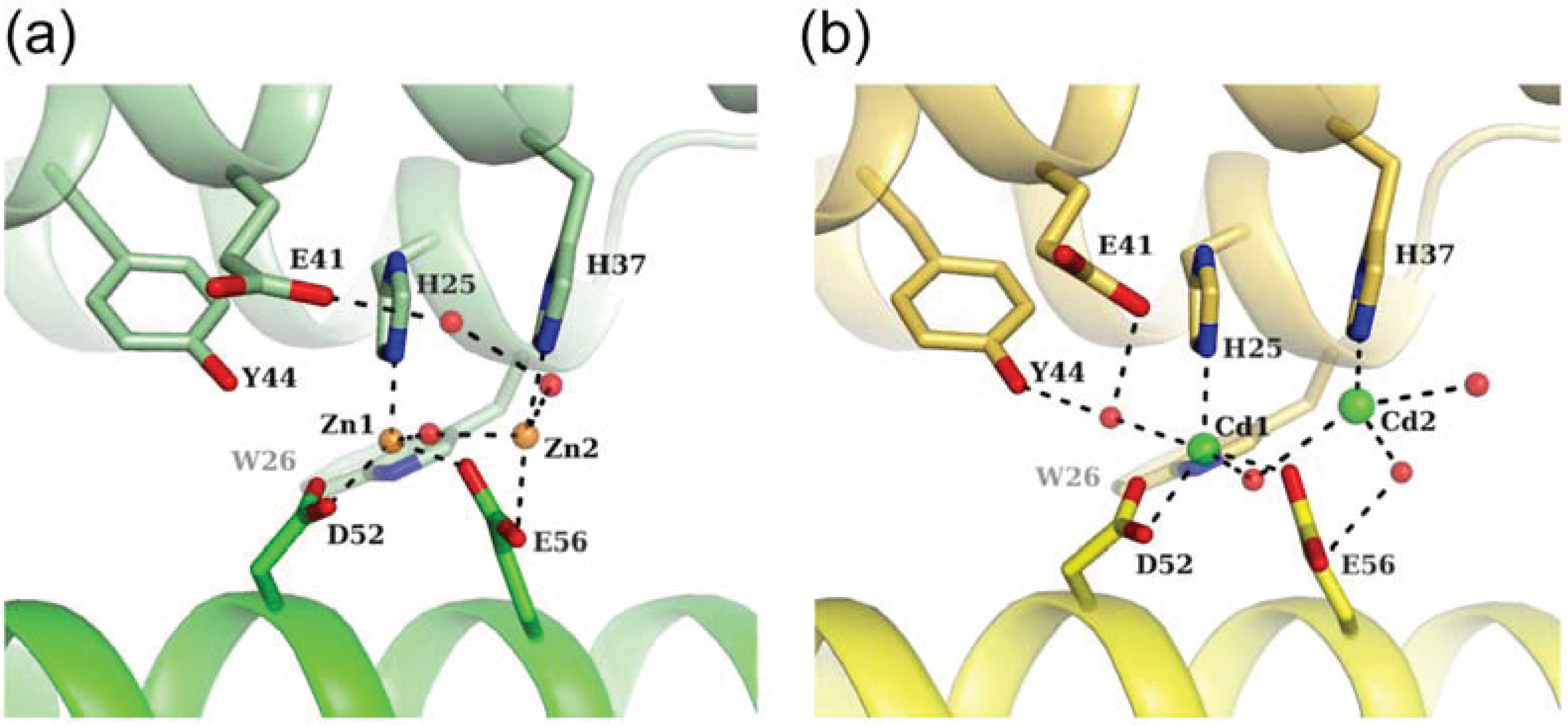
3.2. Zn-Loaded and Cd-Loaded Structures
3.3. Comparison of Metal Coordination
4. Metal Ion Entry through the Three-Fold Pore
4.1. Pore I
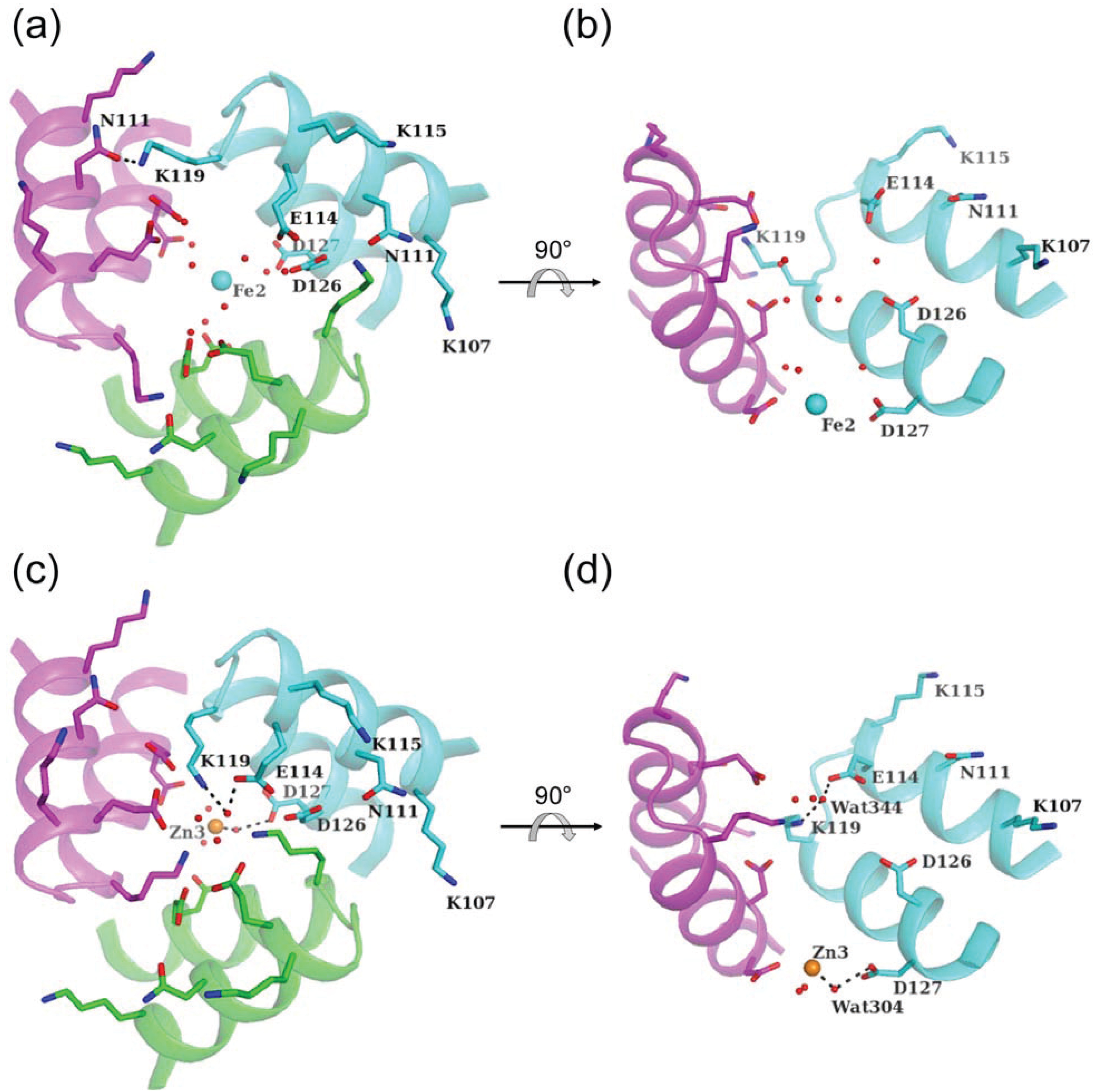
4.2. Pore II
5. Other Metal-Binding Sites
6. Neutrophil Activation

7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Marshall, B.J.; Warren, J.R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984, 1, 1311–1315. [Google Scholar]
- Dunn, B.E.; Cohen, H.; Blaser, M.J. Helicobacter pylori. Clin. Microbiol. Rev. 1997, 10, 720–741. [Google Scholar]
- Brown, L.M. Helicobacter pylori: Epidemiology and routes of transmission. Epidemiol. Rev. 2000, 22, 283–297. [Google Scholar] [CrossRef]
- Goodwin, C.S. Helicobacter pylori gastritis, peptic ulcer, and gastric cancer: Clinical and molecular aspects. Clin. Infect. Dis. 1997, 25, 1017–1019. [Google Scholar]
- Covacci, A.; Telford, J.L.; del Giudice, G.; Parsonnet, J.; Rappuoli, R. Helicobacter pylori virulence and genetic geography. Science 1999, 284, 1328–1333. [Google Scholar]
- Montecucco, C.; Rappuoli, R. Living dangerously: How Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell Biol. 2001, 2, 457–466. [Google Scholar] [CrossRef]
- Montecucco, C.; de Bernard, M. Molecular and cellular mechanisms of action of the vacuolating cytotoxin (VacA) and neutrophil-activating protein (HP-NAP) virulence factors of Helicobacter pylori. Microbes Infect. 2003, 5, 715–721. [Google Scholar] [CrossRef]
- Satin, B.; del Giudice, G.; Bianca, V.D.; Dusi, S.; Laudanna, C.; Tonello, F.; Kelleher, D.; Rappuoli, R.; Montecucco, C.; Rossi, F. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 2000, 191, 1467–1476. [Google Scholar]
- Evans, D.J., Jr.; Evans, D.G.; Takemura, T.; Nakano, H.; Lampert, H.C.; Graham, D.Y.; Granger, D.N.; Kvietys, P.R. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 1995, 63, 2213–2220. [Google Scholar]
- Dundon, W.G.; Nishioka, H.; Polenghi, A.; Papinutto, E.; Zanotti, G.; Montemurro, P.; del Giudice, G.; Rappuoli, R.; Montecucco, C. The neutrophil-activating protein of Helicobacter pylori. Int. J. Med. Microbiol. 2002, 291, 545–550. [Google Scholar]
- Teneberg, S.; Miller-Podraza, H.; Lampert, H.C.; Evans, D.J., Jr.; Evans, D.G.; Danielsson, D.; Karlsson, K.-A. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J. Biol. Chem. 1997, 272, 19067–19071. [Google Scholar] [CrossRef]
- Namavar, F.; Sparrius, M.; Veerman, E.C.; Appelmelk, B.J.; Vandenbroucke-Grauls, C.M. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect. Immun. 1998, 66, 444–447. [Google Scholar]
- Montemurro, P.; Barbuti, G.; Dundon, W.G.; del Giudice, G.; Rappuoli, R.; Colucci, M.; de Rinaldis, P.; Montecucco, C.; Semeraro, N.; Papini, E. Helicobacter pylori neutrophil-activating protein stimulates tissue factor and plasminogen activator inhibitor-2 production by human blood mononuclear cells. J. Infect. Dis. 2001, 183, 1055–1062. [Google Scholar] [CrossRef]
- Polenghi, A.; Bossi, F.; Fischetti, F.; Durigutto, P.; Cabrelle, A.; Tamassia, N.; Cassatella, M.A.; Montecucco, C.; Tedesco, F.; de Bernard, M. The neutrophil-activating protein of Helicobacter pylori crosses endothelia to promote neutrophil adhesion in vivo. J. Immunol. 2007, 178, 1312–1320. [Google Scholar] [CrossRef]
- Montemurro, P.; Nishioka, H.; Dundon, W.G.; de Bernard, M.; del Giudice, G.; Rappuoli, R.; Montecucco, C. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a potent stimulant of mast cells. Eur. J. Immunol. 2002, 32, 671–676. [Google Scholar]
- Amedei, A.; Cappon, A.; Codolo, G.; Cabrelle, A.; Polenghi, A.; Benagiano, M.; Tasca, E.; Azzurri, A.; D’Elios, M.M.; del Prete, G.; et al. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J. Clin. Investig. 2006, 116, 1092–1101. [Google Scholar] [CrossRef]
- D’Elios, M.M.; Amedei, A.; Cappon, A.; del Prete, G.; de Bernard, M. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) as an immune modulating agent. FEMS Immunol. Med. Microbiol. 2007, 50, 157–164. [Google Scholar] [CrossRef]
- De Bernard, M.; D’Elios, M.M. The immune modulating activity of the Helicobacter pylori HP-NAP: Friend or foe? Toxicon 2010, 56, 1186–1192. [Google Scholar] [CrossRef]
- Del Giudice, G.; Covacci, A.; Telford, J.L.; Montecucco, C.; Rappuoli, R. The design of vaccines against Helicobacter pylori and their development. Annu. Rev. Immunol. 2001, 19, 523–563. [Google Scholar] [CrossRef]
- Kabir, S. The current status of Helicobacter pylori vaccines: A review. Helicobacter 2007, 12, 89–102. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Schultze, V.; Rosenkranz, B.; Kaufmann, S.H.E.; Ulrichs, T.; Novicki, D.; Norelli, F.; Contorni, M.; Peppoloni, S.; Berti, D.; et al. Safety and immunogenicity of an intramuscular Helicobacter pylori vaccine in noninfected volunteers: A phase I study. Gastroenterology 2008, 135, 787–795. [Google Scholar]
- Tonello, F.; Dundon, W.G.; Satin, B.; Molinari, M.; Tognon, G.; Grandi, G.; del Giudice, G.; Rappuoli, R.; Montecucco, C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 1999, 34, 238–246. [Google Scholar] [CrossRef]
- Grant, R.A.; Filman, D.J.; Finkel, S.E.; Kolter, R.; Hogle, J.M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 1998, 5, 294–303. [Google Scholar] [CrossRef]
- Ilari, A.; Stefanini, S.; Chiancone, E.; Tsernoglou, D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Biol. 2000, 7, 38–43. [Google Scholar] [CrossRef]
- Wang, G.; Hong, Y.; Olczak, A.; Maier, S.E.; Maier, R.J. Dual roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect. Immun. 2006, 74, 6839–6846. [Google Scholar] [CrossRef]
- Cooksley, C.; Jenks, P.J.; Green, A.; Cockayne, A.; Logan, R.P.H.; Hardie, K.R. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J. Med. Microbiol. 2003, 52, 461–469. [Google Scholar] [CrossRef]
- Wolf, S.G.; Frenkiel, D.; Arad, T.; Finkel, S.E.; Kolter, R.; Minsky, A. DNA protection by stress-induced biocrystallization. Nature 1999, 400, 83–85. [Google Scholar] [CrossRef]
- Haikarainen, T.; Papageorgiou, A.C. Dps-like proteins: Structural and functional insights into a versatile protein family. Cell. Mol. Life Sci. 2010, 67, 341–351. [Google Scholar] [CrossRef]
- Harrison, P.M.; Arosio, P. The ferritins: Molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1996, 1275, 161–203. [Google Scholar] [CrossRef]
- Zanotti, G.; Papinutto, E.; Dundon, W.G.; Battistutta, R.; Seveso, M.; del Giudice, G.; Rappuoli, R.; Montecucco, C. Structure of the neutrophil-activating protein from Helicobacter pylori. J. Mol. Biol. 2002, 323, 125–130. [Google Scholar] [CrossRef]
- Crichton, R.R.; Declercq, J.P. X-ray structures of ferritins and related proteins. Biochim. Biophys. Acta 2010, 1800, 706–718. [Google Scholar] [CrossRef]
- Miyamoto, T.; Asahina, Y.; Miyazaki, S.; Shimizu, H.; Ohto, U.; Noguchi, S.; Satow, Y. Structures of the SEp22 dodecamer, a Dps-like protein from Salmonella enterica subsp. enterica serovar Enteritidis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 17–22. [Google Scholar] [CrossRef]
- Tsuruta, O.; Yokoyama, H.; Fujii, S. A new crystal lattice structure of Helicobacter pylori neutrophil-activating protein (HP-NAP). Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 134–140. [Google Scholar] [CrossRef]
- Yokoyama, H.; Tsuruta, O.; Akao, N.; Fujii, S. Crystal structure of Helicobacter pylori neutrophil-activating protein with a di-nuclear ferroxidase center in a zinc or cadmium-bound form. Biochem. Biophys. Res. Commun. 2012, 422, 745–750. [Google Scholar] [CrossRef]
- Dundas, J.; Ouyang, Z.; Tseng, J.; Binkowski, A.; Turpaz, Y.; Liang, J. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006, 34, W116–W118. [Google Scholar] [CrossRef]
- Ren, B.; Tibbelin, G.; Kajino, T.; Asami, O.; Ladenstein, R. The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J. Mol. Biol. 2003, 329, 467–477. [Google Scholar] [CrossRef]
- Rulíšek, L.; Vondrášek, J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J. Inorg. Biochem. 1998, 71, 115–127. [Google Scholar] [CrossRef]
- Benning, M.M.; Shim, H.; Raushel, F.M.; Holden, H.M. High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta. Biochemistry 2001, 40, 2712–2722. [Google Scholar] [CrossRef]
- Liu, Y.; Ray, W.J., Jr.; Baranidharan, S. Structure of rabbit muscle phosphoglucomutase refined at 2.4 Å resolution. Acta Crystallogr. Sect. D Biol.Crystallogr. 1997, 53, 392–405. [Google Scholar] [CrossRef]
- Holland, D.R.; Hausrath, A.C.; Juers, D.; Matthews, B.W. Structural analysis of zinc substitutions in the active site of thermolysin. ProteinSci. 1995, 4, 1955–1965. [Google Scholar]
- Haikarainen, T.; Tsou, C.C.; Wu, J.J.; Papageorgiou, A.C. Structural characterization and biological implications of di-zinc binding in the ferroxidase center of Streptococcus pyogenes Dpr. Biochem. Biophys. Res. Commun. 2010, 398, 361–365. [Google Scholar] [CrossRef]
- Havukainen, H.; Haataja, S.; Kauko, A.; Pulliainen, A.T.; Salminen, A.; Haikarainen, T.; Finne, J.; Papageorgiou, A.C. Structural basis of the zinc- and terbium-mediated inhibition of ferroxidase activity in Dps ferritin-like proteins. ProteinSci. 2008, 17, 1513–1521. [Google Scholar]
- Papinutto, E.; Dundon, W.G.; Pitulis, N.; Battistutta, R.; Montecucco, C.; Zanotti, G. Structure of two iron-binding proteins from Bacillus anthracis. J. Biol. Chem. 2002, 277, 15093–15098. [Google Scholar]
- Romão, C.V.; Mitchell, E.P.; McSweeney, S. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N terminus. J. Biol. Inorg. Chem. 2006, 11, 891–902. [Google Scholar] [CrossRef]
- Gauss, G.H.; Benas, P.; Wiedenheft, B.; Young, M.; Douglas, T.; Lawrence, C.M. Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry 2006, 45, 10815–10827. [Google Scholar]
- Cuypers, M.G.; Mitchell, E.P.; Romão, C.V.; McSweeney, S.M. The crystal structure of the Dps2 from Deinococcus radiodurans reveals an unusual pore profile with a non-specific metal binding site. J. Mol. Biol. 2007, 371, 787–799. [Google Scholar] [CrossRef]
- Laurence, J.S.; Blanpain, C.; de Leener, A.; Parmentier, M.; LiWang, P.J. Importance of basic residues and quaternary structure in the function of MIP-1β: CCR5 binding and cell surface sugar interactions. Biochemistry 2001, 40, 4990–4999. [Google Scholar] [CrossRef]
- Yang, Y.; Mayo, K.H.; Daly, T.J.; Barry, J.K.; la Rosa, G.J. Subunit association and structural analysis of platelet basic protein and related proteins investigated by 1H-NMR spectroscopy and circular dichroism. J. Biol. Chem. 1994, 269, 20110–20118. [Google Scholar]
- Kottakis, F.; Papadopoulos, G.; Pappa, E.V.; Cordopatis, P.; Pentas, S.; Choli-Papadopoulou, T. Helicobacter pylori neutrophil-activating protein activates neutrophils by its C-terminal region even without dodecamer formation, which is a prerequisite for DNA protection—Novel approaches against Helicobacter pylori inflammation. FEBS J. 2008, 275, 302–317. [Google Scholar] [CrossRef]
- Codolo, G.; Papinutto, E.; Polenghi, A.; D’Elios, M.M.; Zanotti, G.; de Bernard, M. Structure and immunomodulatory property relationship in NapA of Borrelia burgdorferi. Biochim. Biophys. Acta 2010, 1804, 2191–2197. [Google Scholar] [CrossRef]
- Codolo, G.; Amedei, A.; Steere, A.C.; Papinutto, E.; Cappon, A.; Polenghi, A.; Benagiano, M.; Paccani, S.R.; Sambri, V.; del Prete, G.; et al. Borrelia burgdorferi NapA-driven Th17 cell inflammation in lyme arthritis. Arthritis Rheum. 2008, 58, 3609–3617. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yokoyama, H.; Fujii, S. Structures and Metal-Binding Properties of Helicobacter pylori Neutrophil-Activating Protein with a Di-Nuclear Ferroxidase Center. Biomolecules 2014, 4, 600-615. https://doi.org/10.3390/biom4030600
Yokoyama H, Fujii S. Structures and Metal-Binding Properties of Helicobacter pylori Neutrophil-Activating Protein with a Di-Nuclear Ferroxidase Center. Biomolecules. 2014; 4(3):600-615. https://doi.org/10.3390/biom4030600
Chicago/Turabian StyleYokoyama, Hideshi, and Satoshi Fujii. 2014. "Structures and Metal-Binding Properties of Helicobacter pylori Neutrophil-Activating Protein with a Di-Nuclear Ferroxidase Center" Biomolecules 4, no. 3: 600-615. https://doi.org/10.3390/biom4030600



