Particle-Rich Cytoplasmic Structure (PaCS): Identification, Natural History, Role in Cell Biology and Pathology
Abstract
:1. Introduction
2. Types and Function of Ubiquitin-Reactive Cytoplasmic Structures


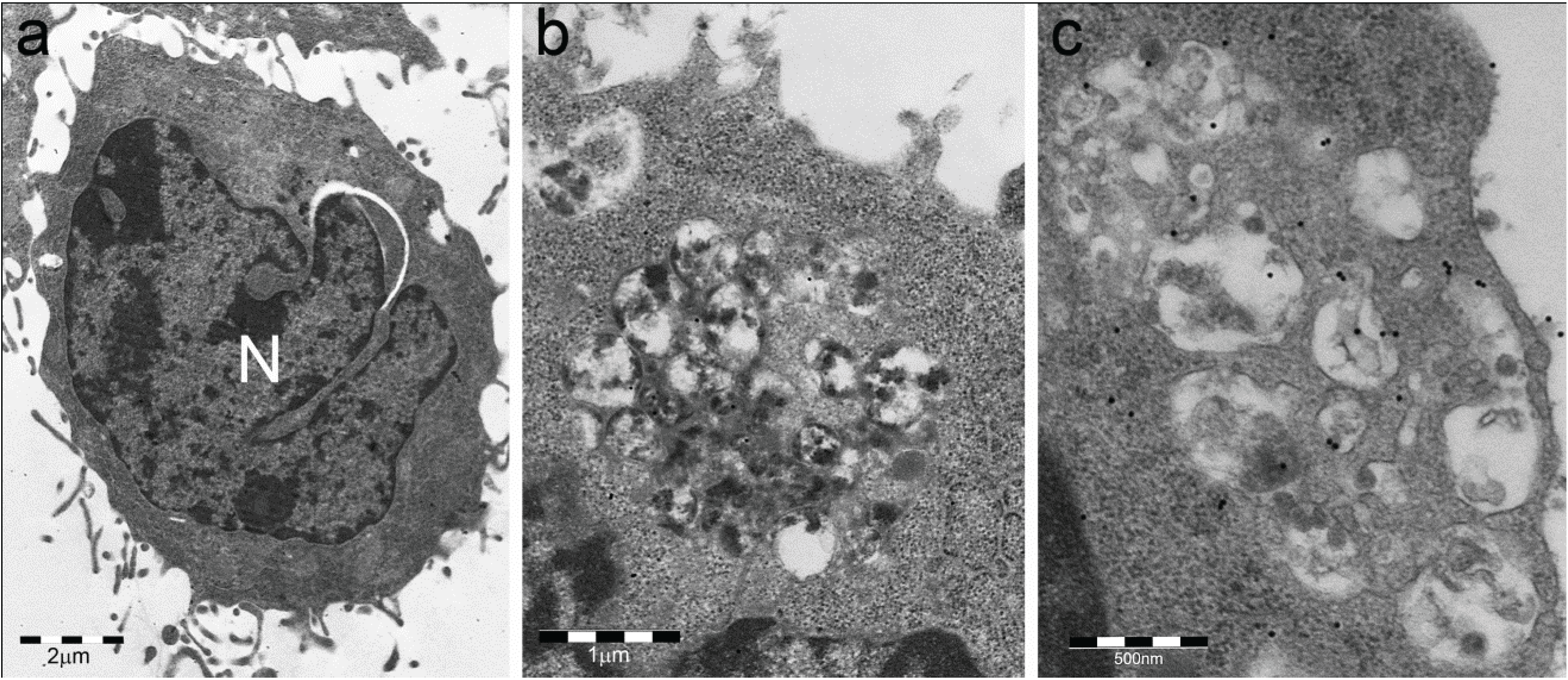
- (a)
- Sequestosomes are reactive for p62 protein (also known as sequestosome 1 or SQSTM1) and ubiquitin and unreactive for the proteasome. They accumulate aggregated insoluble proteins in amorphous to thinly fibrillar (with 5–7 nm-thick fibrils) cytoplasmic bodies mostly devoid of enveloping membranes, though apparently destined to autophagy in a pathway also involving NBR1 (neighbor of BRCA1 gene 1) and ALFY (autophagy-linked FYVE) proteins [6,15,16,17,18].
- (b)
- DALIS [3,4] and ALIS [5] are reactive for ubiquitin and p62 and unreactive for the proteasome. They differ from sequestosomes in their ultrastructure, as recently characterized by Kondylis and coworkers [14] in LPS-activated DCs. Indeed, DALISs contain vesicular membranes and are more or less completely engulfed by LC3-positive double membranes, connected with the late endosomal MHC class II compartment in an “unconventional autophagic pathway” [14].
- (c)
- Pericentriolar aggresomes result from the histone deacetylase 6 (HDAC6)- and dynein-dependent microtubular transport of smaller and peripheral aggregates (often combined with components of the UPS and/or the autophagic-lysosomal pathway) toward the microtubule organizing center (MTOC) [2]. Here, they may associate loosely with each other and form a distinctive “aggregate of aggregates”, rather than a single larger coalescing body [23].
- (d)
- Particle-rich cytoplasmic structures (PaCSs) contain proteasome, polyubiquitinated proteins and glycogen and are characterized ultrastructurally by a collection of barrel-like particles of about 13 nm thick and 13–20 nm (infrequently up to 45) long (Figure 1 and Figure 4; see also [8,15,26]). As PaCSs store highly soluble components poorly preserved by common aldehyde fixatives, their detection requires stronger fixation, such as, for instance, by combined aldehyde and osmium solutions [12,15].
3. Cytochemical and Ultrastructural Characterization of PaCS
4. PaCS Distribution in Cells and Tissues
5. PaCS Origin and Development
6. Intracellular and Extracellular Fate of PaCS
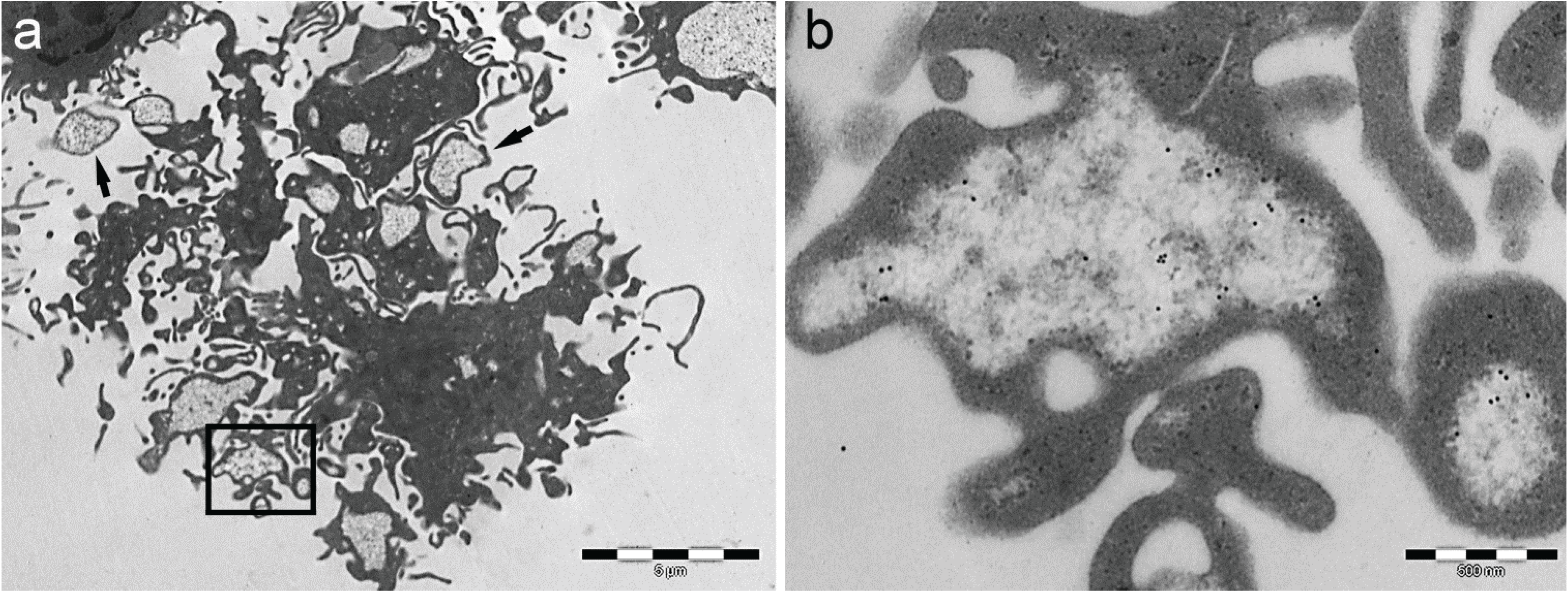
7. Role of PaCS in Pathologic Conditions
7.1. Infectious Diseases

7.2. Neoplastic Growths
8. Conclusions and Perspectives
Abbreviations
| ALFY | autophagy-linked FYVE |
| ALIS | aggresome-like induced structures |
| DALIS | dendritic cell aggresome-like induced structures |
| DCs | dendritic cells |
| GM-CSF | granulocyte macrophage colony stimulating factor |
| HDAC6 | histone deacetylase 6 |
| IL | interleukin |
| IPOD | insoluble protein deposit |
| JUNQ | juxtanuclear quality control |
| LAMP1 | lysosome-associated membrane protein 1 |
| LPS | lipopolysaccharide |
| MHC | major histocompatibility complex |
| MIIC | MHC class II |
| MTOC | microtubule organizing center |
| NBR1 | neighbor of BRCA1 gene 1 |
| NK cells | natural killer cells |
| PaCS | particle-rich cytoplasmic structure |
| RER | rough endoplasmic reticulum |
| SQSTM1 | sequestosome 1 |
| TEM | transmission electron microscopy |
| UPS | ubiquitin proteasome system |
Acknowledgments
Conflicts of Interest
References
- Wójcik, C.; Schroeter, D.; Wilk, S.; Lamprecht, J.; Paweletz, N. Ubiquitin-mediated proteolysis centers in HeLa cells: Indication from studies of an inhibitor of the chymotrypsin-like activity of the proteasome. Eur. J. Cell Biol. 1996, 71, 311–318. [Google Scholar]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- Lelouard, H.; Gatti, E.; Cappello, F.; Gresser, O.; Camosseto, V.; Pierre, P. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature 2002, 417, 177–182. [Google Scholar] [CrossRef]
- Lelouard, H.; Ferrand, V.; Marguet, D.; Bania, J.; Camossetto, V.; David, A.; Gatti, E.; Pierre, P. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 2004, 164, 667–675. [Google Scholar] [CrossRef]
- Szeto, J.; Kaniuk, N.A.; Canadien, V.; Nisman, R.; Mizushima, N.; Yoshimori, T.; Bazett-Jones, D.P.; Brumell, J.H. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy 2006, 2, 189–199. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Øvervatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef]
- Necchi, V.; Sommi, P.; Ricci, V.; Solcia, E. In vivo accumulation of Helicobacter pylori products, NOD1, ubiquitinated proteins and proteasome in a novel cytoplasmic structure. PLoS One 2010, 5, e9716. [Google Scholar] [CrossRef]
- Kuusisto, E.; Salminen, A.; Alafuzoff, I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport 2001, 12, 2085–2090. [Google Scholar] [CrossRef]
- Zatloukal, K.; Stumptner, C.; Fuchsbichler, A.; Heid, H.; Schnoelzer, M.; Kenner, L.; Kleinert, R.; Prinz, M.; Aguzzi, A.; Denk, H. p62 Is a common component of cytoplasmic inclusion in protein aggregation diseases. Am. J. Pathol. 2002, 160, 255–263. [Google Scholar] [CrossRef]
- Willis, M.S.; Patterson, C. Proteotoxicity and cardiac dysfunction—Alzheimer’s disease of the heart? N. Engl. J. Med. 2013, 368, 455–464. [Google Scholar]
- Necchi, V.; Sommi, P.; Vanoli, A.; Manca, R.; Ricci, V.; Solcia, E. Proteasome particle-rich structures are widely present in human epithelial neoplasms: Correlative light, confocal and electron microscopy study. PLoS One 2011, 6, e21317. [Google Scholar] [CrossRef]
- Kaniuk, N.A.; Lam, G.Y.; Ma, C.; Galindo-Mata, E.; Jones, N.; Vallance, B.A.; Brumell, J.H. Citrobacterrodentium infection induces MyD88-dependent formation of ubiquitinated protein aggregates in the intestinal epithelium. J. Innate Immun. 2011, 3, 83–98. [Google Scholar] [CrossRef]
- Kondylis, V.; van Nispen Tot Pannerden, H.E.; van Dijk, S.; Ten Broeke, T.; Wubbolts, R.; Geerts, W.J.; Seinen, C.; Mutis, T.; Heijnen, H.F. Endosome-mediated autophagy: An unconventional MIIC-driven autophagic pathway operational in dendritic cells. Autophagy 2013, 9, 861–880. [Google Scholar] [CrossRef]
- Sommi, P.; Necchi, V.; Vitali, A.; Montagna, D.; de Luigi, A.; Salmona, M.; Ricci, V.; Solcia, E. PaCSis a novel cytoplasmic structure containing functional proteasome and inducible by cytokines/trophic factors. PLoS One 2013, 8, e82560. [Google Scholar] [CrossRef]
- Denk, H.; Stumptner, C.; Fuchsbichler, A.; Müller, T.; Farr, G.; Müller, W.; Terracciano, L.; Zatloukal, K. Are the Mallory bodies and intracellular hyaline bodies in neoplastic and non-neoplastic hepatocytes related? J. Pathol. 2006, 208, 653–661. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Fujimuro, M.; Sawada, H.; Yokosawa, H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994, 349, 173–180. [Google Scholar] [CrossRef]
- Iwata, A.; Riley, B.E.; Johnston, J.A.; Kopito, R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005, 280, 40282–40292. [Google Scholar] [CrossRef]
- Wigley, W.C.; Fabunmi, R.P.; Lee, M.G.; Marino, C.R.; Muallem, S.; DeMartino, G.N.; Thomas, P.J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999, 145, 481–490. [Google Scholar] [CrossRef]
- Waelter, S.; Boeddrich, A.; Lurz, R.; Scherzinger, E.; Lueder, G.; Lehrach, H.; Wanker, E.E. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 2001, 12, 1393–1407. [Google Scholar] [CrossRef]
- García-Mata, R.; Bebök, Z.; Sorscher, E.J.; Sztul, E.S. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1999, 146, 1239–1254. [Google Scholar]
- Canadien, V.; Tan, T.; Zilber, R.; Szeto, J.; Perrin, A.J.; Brumell, J.H. Cutting edge: Microbial products elicit formation of dendritic cell aggresome-like induced structures in macrophages. J. Immunol. 2005, 174, 2471–2475. [Google Scholar] [CrossRef]
- Pierre, P. Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol. Rev. 2005, 207, 184–190. [Google Scholar] [CrossRef]
- Necchi, V.; Sommi, P.; Vitali, A.; Vanoli, A.; Savoia, A.; Ricci, V.; Solcia, E. Polyubiquitinated proteins, proteasome, and glycogen characterize the particle-rich cytoplasmic structure (PaCS) of neoplastic and fetal cells. Histochem. Cell Biol. 2014, 141, 483–497. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Li, L.; Chin, L.S. Aggresome formation and neurodegenerative diseases: Therapeutic implications. Curr. Med. Chem. 2008, 15, 47–60. [Google Scholar]
- Necchi, V.; Minelli, A.; Sommi, P.; Vitali, A.; Caruso, R.; Longoni, D.; Frau, M.R.; Nasi, C.; de Gregorio, F.; Zecca, M.; et al. Ubiquitin-proteasome-rich cytoplasmic structures in neutrophils of patients with Shwachman-Diamond syndrome. Haematologica 2012, 97, 1057–1063. [Google Scholar] [CrossRef]
- Necchi, V.; Balduini, A.; Noris, P.; Barozzi, S.; Sommi, P.; di Buduo, C.; Balduini, C.L.; Solcia, E.; Pecci, A. Ubiquitin/proteasome-rich particulate cytoplasmic structures (PaCSs) in the platelets and megakaryocytes of ANKRD26-related thrombo-cytopenia. Thromb. Haemost. 2013, 109, 263–271. [Google Scholar]
- Kanayama, H.; Tanaka, K.; Aki, M.; Kagawa, S.; Miyaji, H.; Satoh, M.; Okada, F.; Sato, S.; Shimbara, N.; Ichihara, A. Changes in expressions of proteasome and ubiquitin genes in human renal cancer cells. Cancer Res. 1991, 51, 6677–6685. [Google Scholar]
- Lavabre-Bertrand, T.; Henry, L.; Carillo, S.; Guiraud, I.; Ouali, A.; Dutaud, D.; Aubry, L.; Rossi, J.F.; Bureau, J.P. Plasma proteasome level is a potential marker in patients with solid tumors and hemopoietic malignancies. Cancer 2001, 92, 2493–2500. [Google Scholar] [CrossRef]
- Bazzaro, M.; Lee, M.K.; Zoso, A.; Stirling, W.L.; Santillan, A.; Shih, IeM.; Roden, R.B. Ubiquitin-proteasome system stress sensitizes ovarian cancer to proteasome inhibitor-induced apoptosis. Cancer Res. 2006, 66, 3754–3763. [Google Scholar]
- Ryu, K.-Y.; Maehr, R.; Gilchrist, C.A.; Long, M.A.; Bouley, D.M.; Mueller, B.; Ploegh, H.L.; Kopito, R.R. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. EMBO J. 2007, 26, 2693–2706. [Google Scholar] [CrossRef]
- Richardson, P.G.; Mitsiades, C.; Hideshima, T.; Anderson, K.C. Proteasome inhibition in the treatment of cancer. Cell Cycle 2005, 4, 290–296. [Google Scholar]
- Baumeister, W.; Walz, J.; Zühl, F.; Seemüller, E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 1998, 92, 367–380. [Google Scholar] [CrossRef]
- Hirano, Y.; Hendil, K.B.; Yashiroda, H.; Iemura, S.; Nagane, R.; Hioki, Y.; Natsume, T.; Tanaka, K.; Murata, S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 2005, 437, 1381–1385. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Liu, C.-W.; Corboy, M.J.; DeMartino, G.N.; Thomas, P.J. Endoproteolytic activity of the proteasome. Science 2003, 299, 408–411. [Google Scholar] [CrossRef]
- Baugh, J.M.; Viktorova, E.G.; Pilipenko, E.V. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009, 386, 814–827. [Google Scholar] [CrossRef]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermark, G.; Grune, T.; Davies, K.J. The immunoproteasome, the 20S proteasome, and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–594. [Google Scholar] [CrossRef]
- Bendayan, M.; Londono, I.; Kemp, B.E.; Hardie, G.D.; Ruderman, N.; Prentki, M. Association of AMP-activated protein kinase subunits with glycogen particles as revealed in situ by immunoelectron microscopy. J. Histochem. Cytochem. 2009, 57, 963–971. [Google Scholar] [CrossRef]
- Kloetzel, P.M. The proteasome and MHC class I antigen processing. Biochim. Biophys. Acta 2004, 1695, 225–233. [Google Scholar]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Egerer, K.; Kuckelkorn, U.; Rudolph, P.E.; Rückert, J.C.; Dörner, T.; Burmester, G.R.; Kloetzel, P.M.; Feist, E. Circulating proteasomes are markers of cell damage and immunologic activity in autoimmune diseases. J. Rheumatol. 2002, 29, 2045–2052. [Google Scholar]
- Sixt, S.U.; Dahlmann, B. Extracellular, circulating proteasomes and ubiquitin—Incidence and relevance. Biochim. Biophys. Acta 2008, 1782, 817–823. [Google Scholar]
- Necchi, V.; Candusso, M.E.; Tava, F.; Luinetti, O.; Ventura, U.; Fiocca, R.; Ricci, V.; Solcia, E. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology 2007, 132, 1009–1023. [Google Scholar] [CrossRef]
- Terebiznik, M.R.; Raju, D.; Vázquez, C.L.; Torbricki, K.; Kulkarni, R.; Blanke, S.R.; Yoshimori, T.; Colombo, M.I.; Jones, N.L. Effect of Helicobacter pylori’s vacuolatingcytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy 2009, 5, 370–379. [Google Scholar] [CrossRef]
- Zheng, Y.T.; Shahnazari, S.; Brech, A.; Lamark, T.; Johansen, T.; Brumell, J.H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009, 183, 5909–5916. [Google Scholar] [CrossRef]
- Kwak, S.; Masaki, T.; Ishiura, S.; Sugita, H. Multicatalytic proteinase is present in Lewy bodies and neurofibrillary tangles in diffuse Lewy body disease brains. Neurosci. Lett. 1991, 128, 21–24. [Google Scholar] [CrossRef]
- McNaught, K.S.; Björklund, L.M.; Belizaire, R.; Isacson, O.; Jenner, P.; Olanow, C.W. Proteasome inhibition causes nigral degeneration with inclusion bodies in rats. Neuroreport 2002, 13, 1437–1441. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Solcia, E.; Sommi, P.; Necchi, V.; Vitali, A.; Manca, R.; Ricci, V. Particle-Rich Cytoplasmic Structure (PaCS): Identification, Natural History, Role in Cell Biology and Pathology. Biomolecules 2014, 4, 848-861. https://doi.org/10.3390/biom4030848
Solcia E, Sommi P, Necchi V, Vitali A, Manca R, Ricci V. Particle-Rich Cytoplasmic Structure (PaCS): Identification, Natural History, Role in Cell Biology and Pathology. Biomolecules. 2014; 4(3):848-861. https://doi.org/10.3390/biom4030848
Chicago/Turabian StyleSolcia, Enrico, Patrizia Sommi, Vittorio Necchi, Agostina Vitali, Rachele Manca, and Vittorio Ricci. 2014. "Particle-Rich Cytoplasmic Structure (PaCS): Identification, Natural History, Role in Cell Biology and Pathology" Biomolecules 4, no. 3: 848-861. https://doi.org/10.3390/biom4030848



