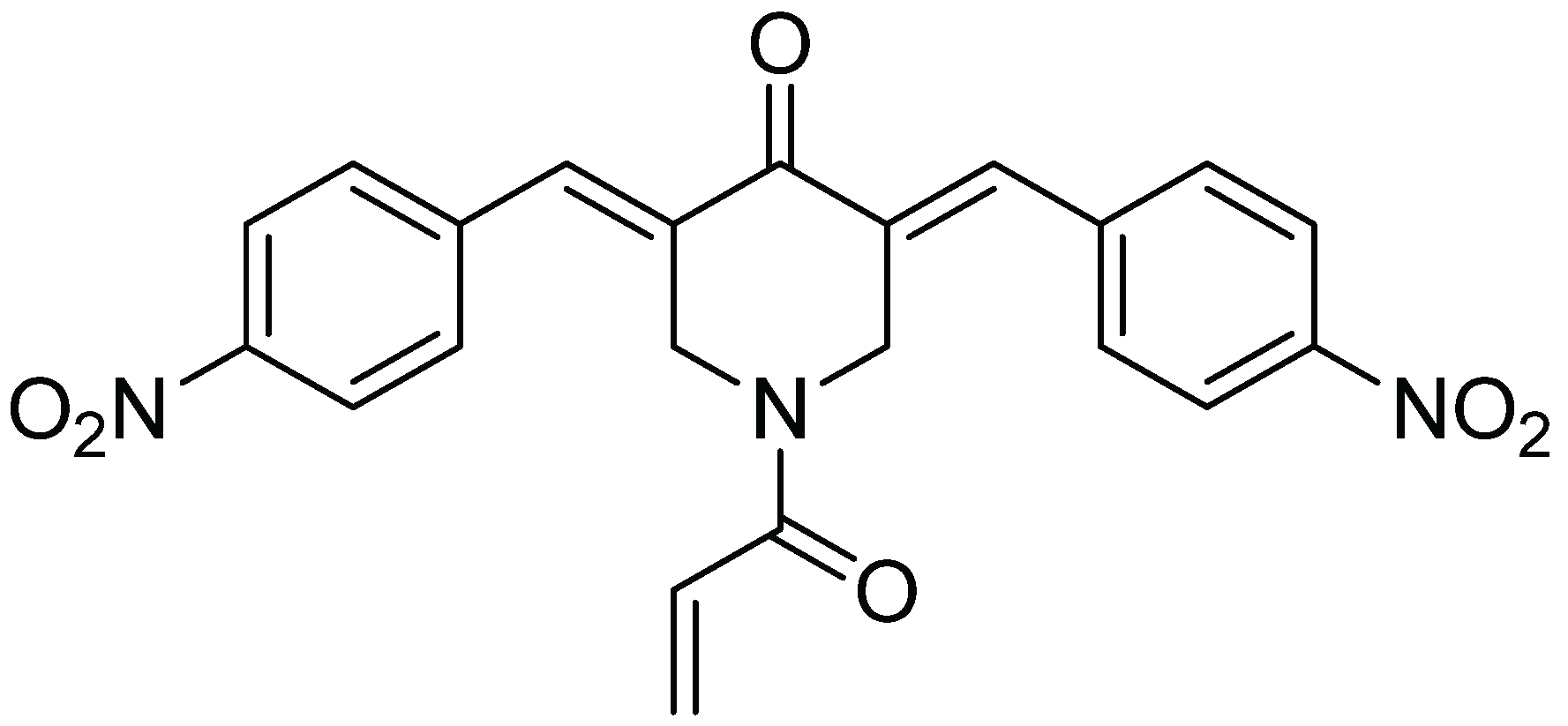
Inhibitory Effect of b-AP15 on the 20S Proteasome
Abstract
:1. Introduction
2. Results and Discussion
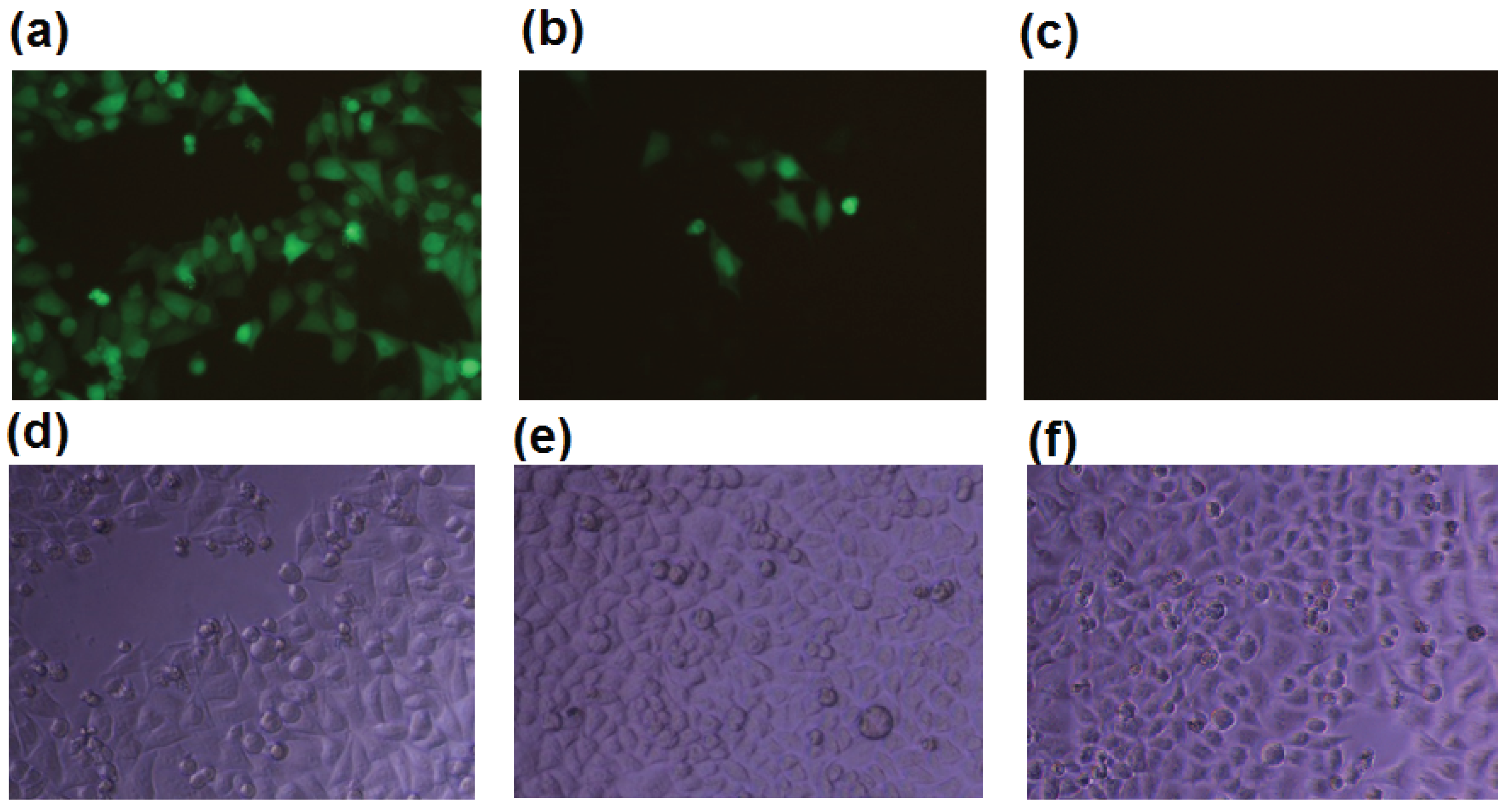
2.1. b-AP15 Potently Inhibited the Proteasome in a Cell-Based Assay
2.2. b-AP15 Inhibited Both 19S Regulatory Particle and the 20S Proteasome

| IC50 (μM) | |||
|---|---|---|---|
| Compound | PA28 | PALAME | SDS |
| b-AP15 | 18.1 ± 3.22 | 26.3 ± 3.15 | >48 |
| >48 b | |||
| 40.6 ± 5.23 c | |||
| Lactacystin | 5.8 ± 0.76 | 5.3 ± 0.68 | 6.4 ± 0.71 |
| Bortezomib | 0.0078 ±0.0012 | 0.0065 ±0.00093 | 0.0087 ±0.00015 |
3. Experimental
3.1. Cell-Based Proteasome Inhibition Assay
3.2. 19S Regulatory Particle De-Ubiquitinase Assay
3.3. 20S Proteasome Assay
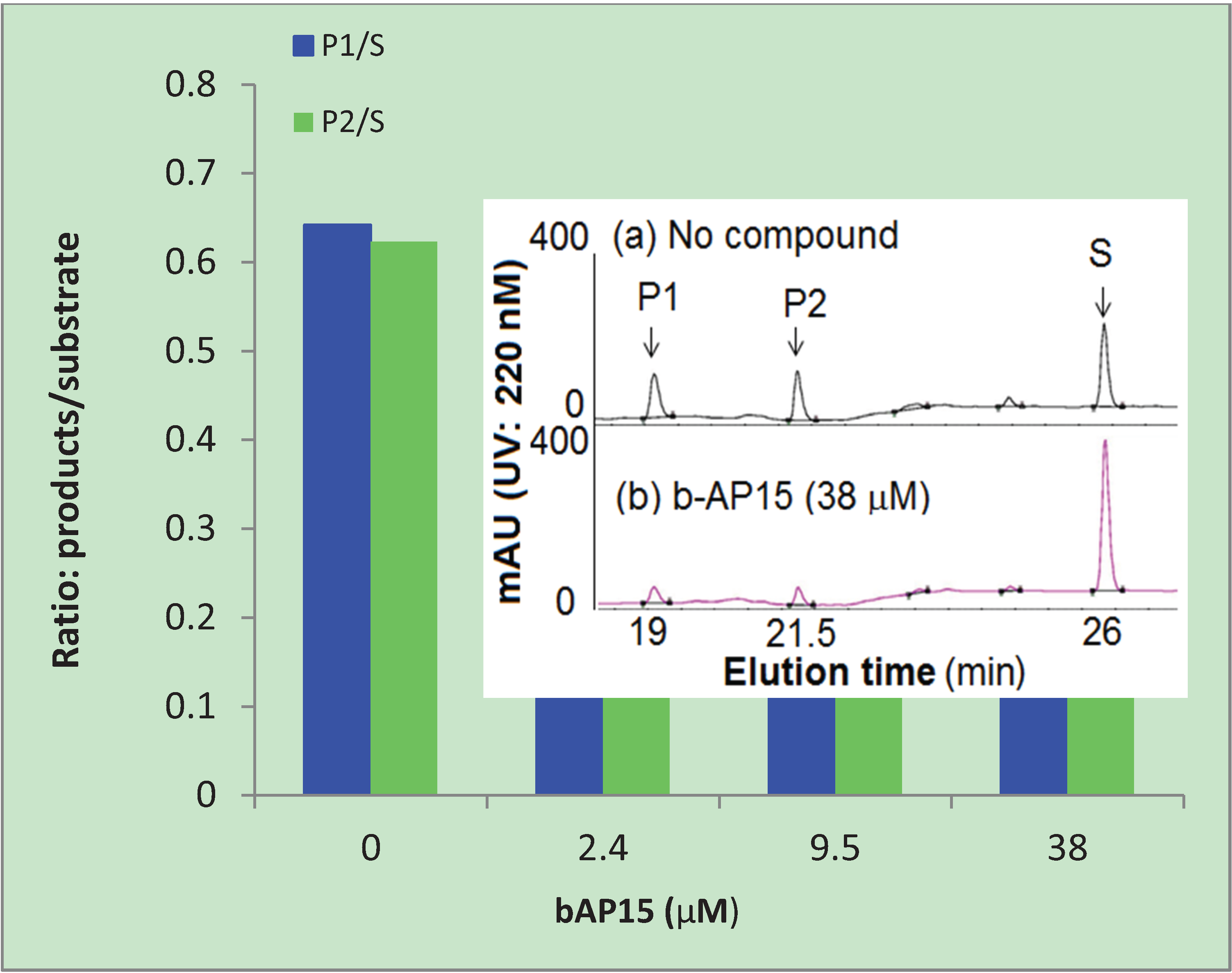
3.4. HPLC Analysis of the Substrates and Products of the 20S Proteasome
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wong, E.; Cuervo, A.M. Integration of clearance mechanisms: The proteasome and autophagy. Cold Spring Harb. Perspect. Biol. 2010. [Google Scholar] [CrossRef]
- Kish-Trier, E.; Hill, C.P. Structural biology of the proteasome. Annu. Rev. Biophys. 2013, 42, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Aso, E.; Lomoio, S.; López-González, I.; Joda, L.; Carmona, M.; Fernández-Yagüe, N.; Moreno, J.; Juvés, S.; Pujol, A.; Pamplona, R.; et al. Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer’s disease. Brain Pathol. 2012, 22, 636–653. [Google Scholar]
- Seo, H.; Sonntag, K.C.; Kim, W.; Cattaneo, E.; Isacson, O. Proteasome activator enhances survival of Huntington’s disease neuronal model cells. PLoS One 2007, 2, e238. [Google Scholar] [CrossRef]
- Kloetzel, P.M.; Ossendorp, F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 2004, 16, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.; Kim, K.B.; Crews, C.M. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 2001, 21, 245–273. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; van der Linden, W.A.; Overkleeft, H.S. Proteasome inhibitors: An expanding army attacking a unique target. Chem. Biol. 2012, 19, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Gräwert, M.A.; Groll, M. Exploiting nature’s rich source of proteasome inhibitors as starting points in drug development. Chem. Commun. 2012, 48, 1364–1378. [Google Scholar] [CrossRef]
- Stein, M.L.; Groll, M. Applied techniques for mining natural proteasome inhibitors. Biochim. Biophys. Acta 2014, 1843, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.; Kauffman, M. Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 2004, 22, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Lawasut, P.; Chauhan, D.; Laubach, J.; Hayes, C.; Fabre, C.; Maglio, M.; Mitsiades, C.; Hideshima, T.; Anderson, K.C.; Richardson, P.G. New proteasome inhibitors in myeloma. Curr. Hematol. Malig. Rep. 2012, 7, 258–266. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, P.; Brnjic, S.; Olofsson, M.H.; Fryknäs, M.; Lindsten, K.; de Cesare, M.; Perego, P.; Sadeghi, B.; Hassan, M.; Larsson, R.; et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011, 17, 1636–1640. [Google Scholar]
- Tian, Z.; D’Arcy, P.; Wang, X.; Ray, A.; Tai, Y.T.; Hu, Y.; Carrasco, R.D.; Richardson, P.; Linder, S.; Chauhan, D.; et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance. Blood 2014, 123, 706–716. [Google Scholar]
- Menéndez-Bnito, V.; Heessen, S.; Dantuma, N.P. Monitoring of ubiquitin-dependent proteolysis with green fluorescent protein substrates. Methods Enzymol. 2005, 399, 490–511. [Google Scholar] [PubMed]
- Dang, Z.; Jung, K.; Qian, K.D.; Lee, K.H.; Huang, L.; Chen, C.H. Synthesis of lithocholic acid derivatives as proteasome regulators. ACS Med. Chem. Lett. 2012, 3, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yu, D.; Ho, P.; Qian, K.D.; Lee, K.H.; Chen, C.H. Synthesis and proteasome inhibition of glycyrrhetinic acid derivatives. Bioorg. Med. Chem. 2008, 16, 6696–6701. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, A.A.; Navon, A.; Groll, M.; Smith, D.M.; Reis, C.; Goldberg, A.L. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J. Biol. Chem. 2007, 282, 22921–22929. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.P.; Masters, E.I.; Whitby, F.G. The 11S regulators of 20S proteasome activity. Curr. Top. Microbiol. Immunol. 2002, 268, 73–89. [Google Scholar] [PubMed]
- Rechsteiner, M.; Hill, C.P. Mobilizing the proteolytic machine: Cell biological roles of proteasome activators and inhibitors. Trends Cell. Biol. 2005, 15, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Stafford, W.; Mazurkiewicz, M.; Fryknäs, M.; Brjnic, S.; Zhang, X.; Gullbo, J.; Larsson, R.; Arnér, E.S.; D’Arcy, P.; et al. The 19S Deubiquitinase inhibitor b-AP15 is enriched in cells and elicits rapid commitment to cell death. Mol. Pharmacol. 2014, 85, 932–945. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, L.; Jung, K.; Chen, C.H. Inhibitory Effect of b-AP15 on the 20S Proteasome. Biomolecules 2014, 4, 931-939. https://doi.org/10.3390/biom4040931
Huang L, Jung K, Chen CH. Inhibitory Effect of b-AP15 on the 20S Proteasome. Biomolecules. 2014; 4(4):931-939. https://doi.org/10.3390/biom4040931
Chicago/Turabian StyleHuang, Li, Katherine Jung, and Chin Ho Chen. 2014. "Inhibitory Effect of b-AP15 on the 20S Proteasome" Biomolecules 4, no. 4: 931-939. https://doi.org/10.3390/biom4040931




