Nuclear Transport of Yeast Proteasomes
Abstract
:1. Where Are Proteins Degraded by Proteasomes?
2. What Do We Know about Proteasome Assembly to Understand Proteasome Dynamics?
3. Do Parallels Exist between Nuclear Transport of Proteasomes and Other Macromolecular Machineries?
4. What Do We Know about the Basic Concept of Nuclear Transport through Nuclear Pores?
5. How Are Yeast Proteasomes Imported into the Nucleus during Cell Division?
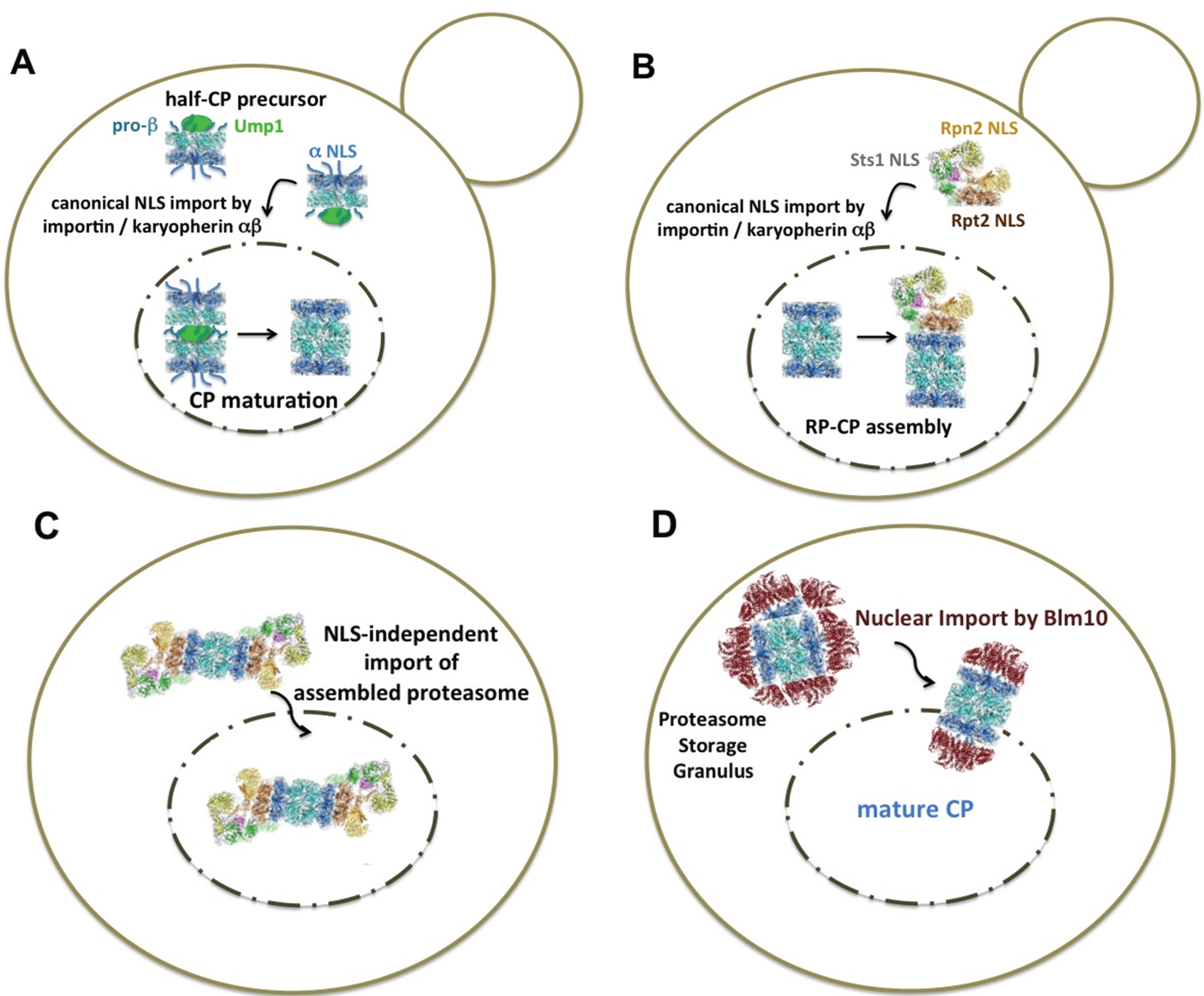
| Structure | Name and Specification |
|---|---|
 | Ump1-associated CP precursor complex, symbolized as half-CP. Five of seven β subunits (cyan) are synthesized with propeptides [16,17]. Four of seven α subunits (blue) carry classical NLS (α1, α2, α4 and α6 in yeast) [39,40,41]. The α ring is disordered and can bind to Blm10 and the canonical NLS receptor importin/karyopherin αβ [35,49]. |
 | In the pre-holo-CP or nascent CP, the β subunit propeptides are processed and Ump1 is degraded [17]. The α ring is disordered and can bind to Blm10 and possibly to the NLS receptor, importin/karyopherin αβ [49]. |
 | The interior between the β rings (cyan) of the mature CP harbors the proteolytic sites [18]. The α rings (blue) are closed and do not bind to Blm10 or the NLS receptor, importin/karyopherin αβ [20,35,51]. |
 | Mature CP bound to Blm10 (red) represents the nuclear import receptor-cargo complex upon exit from quiescence, when CP precursor complexes are unavailable [48]. The α rings (blue) bound to Blm10 are disordered [52]. |
6. Do Multiple Import Pathways Exist for Dividing Yeast Cells? The Canonical Import Pathway of Proteasomal Modules vs. the Non-Canonical Import Pathway of Holo-Enzyme Complexes
7. Is the Concept of Nuclear Import of Yeast Proteasomes Applicable to Higher Eukaryotic Cells?
8. How Are Yeast Proteasomes Imported into the Nucleus upon the Exit from Quiescence?
9. Do Alternative Import Pathways of Proteasomes Exist in Quiescent Vertebrate Cells?
10. Is There a Common Principle behind the Different Import Pathways of Proteasomes?
11. What Is Known about Nuclear Export of Yeast Proteasomes?
12. Conclusions
Acknowledgment
Conflicts of Interest
References
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [PubMed]
- Kirschner, M. Intracellular proteolysis. Trends Cell Biol. 1999, 9, M42–M45. [Google Scholar] [CrossRef] [PubMed]
- Von Mikecz, A. The nuclear ubiquitin-proteasome system. J. Cell Sci. 2006, 119, 1977–1984. [Google Scholar] [CrossRef] [PubMed]
- Vabulas, R.M.; Hartl, F.U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 2005, 310, 1960–1963. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, P.S.; Oeser, M.L.; Abraham, A.C.; Kaganovich, D.; Gardner, R.G. Cellular maintenance of nuclear protein homeostasis. Cell. Mol. Life Sci. 2013, 71, 1865–1879. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Madura, K. Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics 2014, 197, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Arai, N.; Tanaka, K.; Saeki, Y. Cytoplasmic proteasomes are not indispensable for cell growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2013, 436, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.W.; Ju, D.; Xie, Y. Nuclear import factor Srp1 and its associated protein Sts1 couple ribosome-bound nascent polypeptides to proteasomes for cotranslational degradation. J. Biol. Chem. 2014, 289, 2701–2710. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef] [PubMed]
- Enenkel, C. Proteasome dynamics. Biochim. Biophys. Acta 2013, 1843, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Hugle, B.; Kleinschmidt, J.A.; Franke, W.W. The 22S cylinder particles of Xenopus laevis. II. Immunological characterization and localization of their proteins in tissues and cultured cells. Eur. J. Cell Biol. 1983, 32, 157–163. [Google Scholar] [PubMed]
- Kleinschmidt, J.A.; Hugle, B.; Grund, C.; Franke, W.W. The 22S cylinder particles of Xenopus laevis. I. Biochemical and electron microscopic characterization. Eur. J. Cell Biol. 1983, 32, 143–156. [Google Scholar] [PubMed]
- Wojcik, C.; DeMartino, G.N. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 2003, 35, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, W.; Walz, J.; Zuhl, F.; Seemuller, E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 1998, 92, 367–380. [Google Scholar] [PubMed]
- Enenkel, C. Using native gel electrophoresis and phosphofluoroimaging to analyze GFP-tagged proteasomes. In Ubiquitin Family Modifiers and the Proteasome: Reviews and Protocols; Dohmen, R.J., Scheffner, M., Eds.; Humana Press: New York, NY, USA, 2011; pp. 339–348. [Google Scholar]
- Chen, P.; Hochstrasser, M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 1996, 86, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Ramos, P.C.; Hockendorff, J.; Johnson, E.S.; Varshavsky, A.; Dohmen, R.J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 1998, 92, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Le Tallec, B.; Barrault, M.B.; Courbeyrette, R.; Guerois, R.; Marsolier-Kergoat, M.C.; Peyroche, A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell 2007, 27, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Mol. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Liu, C.W.; Corboy, M.J.; DeMartino, G.N.; Thomas, P.J. Endoproteolytic activity of the proteasome. Science 2003, 299, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Besche, H.C.; Peth, A.; Goldberg, A.L. Getting to first base in proteasome assembly. Cell 2009, 138, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Rubin, D.M.; Fried, V.A.; Finley, D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998, 18, 3149–3162. [Google Scholar] [PubMed]
- Beck, F.; Unverdorben, P.; Bohn, S.; Schweitzer, A.; Pfeifer, G.; Sakata, E.; Nickell, S.; Plitzko, J.M.; Villa, E.; Baumeister, W.; et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl. Acad. Sci. USA 2012, 109, 14870–14875. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Bronner, V.; Zhang, D.; Fushman, D.; Glickman, M.H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 2012, 287, 14659–14671. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Nissan, T.A.; Bassler, J.; Petfalski, E.; Tollervey, D.; Hurt, E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002, 21, 5539–5547. [Google Scholar] [CrossRef] [PubMed]
- Hulsmann, B.B.; Labokha, A.A.; Gorlich, D. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 2012, 150, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B.; Aebi, U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 2003, 4, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Lemke, E.A. Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153. Biophys. J. 2011, 101, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Wente, S.R.; Rout, M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010. [Google Scholar] [CrossRef]
- Enenkel, C.; Blobel, G.; Rexach, M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995, 270, 16499–16502. [Google Scholar] [PubMed]
- Pante, N.; Kann, M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Wendler, P.; Lehmann, A.; Janek, K.; Baumgart, S.; Enenkel, C. The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin ab and proteasome functions. J. Biol. Chem. 2004, 279, 37751–37762. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.; Janek, K.; Braun, B.; Kloetzel, P.M.; Enenkel, C. 20S proteasomes are imported as precursor complexes into the nucleus of yeast. J. Mol. Biol. 2002, 317, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Isono, E.; Nishihara, K.; Saeki, Y.; Yashiroda, H.; Kamata, N.; Ge, L.; Ueda, T.; Kikuchi, Y.; Tanaka, K.; Nakano, A.; et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell 2007, 18, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Savulescu, A.F.; Rotem, A.; Harel, A. Proteasomes crossing the nuclear border. Nucleus 2011, 2, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Romero, L.; Chuang, S.M.; Tournier, V.; Joshi, K.K.; Lee, J.A.; Kovvali, G.; Madura, K. Sts1 plays a key role in targeting proteasomes to the nucleus. J. Biol. Chem. 2011, 286, 3104–3118. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.R.; Kania, M.; Baumeister, W.; Nederlof, P.M. Import of human and thermoplasma 20S proteasomes into nuclei of Hela cells requires functional NLS sequences. Eur. J. Cell Biol. 1997, 73, 105–113. [Google Scholar] [PubMed]
- Nederlof, P.M.; Wang, H.R.; Baumeister, W. Nuclear localization signals of human and thermoplasma proteasomal alpha subunits are functional in vitro. Proc. Natl. Acad. Sci. USA 1995, 92, 12060–12064. [Google Scholar] [CrossRef] [PubMed]
- Knuehl, C.; Seelig, A.; Brecht, B.; Henklein, P.; Kloetzel, P.M. Functional analysis of eukaryotic 20S proteasome nuclear localization signal. Exp. Cell Res. 1996, 225, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Tone, Y.; Toh-e, E.A. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev. 2002, 16, 3142–3157. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.Y.; Chen, R.H. Cdc48 chaperone and adaptor Ubx4 distribute the proteasome in the nucleus for anaphase proteolysis. J. Biol. Chem. 2013, 288, 37180–37191. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Yoshimura, T.; Tamura, T.; Fujiwara, T.; Kumatori, A.; Ichihara, A. Possible mechanism of nuclear translocation of proteasomes. FEBS Lett. 1990, 271, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Mayr, J.; Wang, H.R.; Nederlof, P.; Baumeister, W. The import pathway of human and Thermoplasma 20S proteasomes into Hela cell nuclei is different from that of classical NLS-bearing proteins. Biol. Chem. 1999, 380, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Savulescu, A.F.; Shorer, H.; Kleifeld, O.; Cohen, I.; Gruber, R.; Glickman, M.H.; Harel, A. Nuclear import of an intact preassembled proteasome particle. Mol. Biol. Cell 2011, 22, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Pack, C.G.; Yukii, H.; Toh, E.A.; Kudo, T.; Tsuchiya, H.; Kaiho, A.; Sakata, E.; Murata, S.; Yokosawa, H.; Sako, Y.; et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 2014. [Google Scholar] [CrossRef]
- Weberruss, M.H.; Savulescu, A.F.; Jando, J.; Bissinger, T.; Harel, A.; Glickman, M.H.; Enenkel, C. Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 2013, 32, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Fehlker, M.; Wendler, P.; Lehmann, A.; Enenkel, C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003, 4, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, P.C.; He, J.; Morris, E.P. Molecular model of the human 26S proteasome. Mol. Cell 2012, 46, 54–66. [Google Scholar]
- Lehmann, A.; Jechow, K.; Enenkel, C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008, 9, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Sadre-Bazzaz, K.; Whitby, F.G.; Robinson, H.; Formosa, T.; Hill, C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 2010, 37, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kusmierczyk, A.R.; Wong, P.; Emili, A.; Hochstrasser, M. Beta-subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007, 26, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A.; Niewienda, A.; Jechow, K.; Janek, K.; Enenkel, C. Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell 2010, 38, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Tabb, M.M.; Tongaonkar, P.; Vu, L.; Nomura, M. Evidence for separable functions of Srp1p, the yeast homolog of importin alpha (karyopherin alpha): Role for Srp1p and Sts1p in protein degradation. Mol. Cell. Biol. 2000, 20, 6062–6073. [Google Scholar] [CrossRef] [PubMed]
- Tatebe, H.; Yanagida, M. Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr. Biol. 2000, 10, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Tonthat, N.K.; Glover, T.; Xu, W.; Koonin, E.V.; Yanagida, M.; Schumacher, M.A. Implications for proteasome nuclear localization revealed by the structure of the nuclear proteasome tether protein cut8. Proc. Natl. Acad. Sci. USA 2011, 108, 16950–16955. [Google Scholar] [CrossRef] [PubMed]
- Amsterdam, A.; Pitzer, F.; Baumeister, W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc. Natl. Acad. Sci. USA 1993, 90, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Reits, E.A.; Benham, A.M.; Plougastel, B.; Neefjes, J.; Trowsdale, J. Dynamics of proteasome distribution in living cells. EMBO J. 1997, 16, 6087–6094. [Google Scholar] [CrossRef] [PubMed]
- Kremer, M.; Henn, A.; Kolb, C.; Basler, M.; Moebius, J.; Guillaume, B.; Leist, M.; van den Eynde, B.J.; Groettrup, M. Reduced immunoproteasome formation and accumulation of immunoproteasomal precursors in the brains of lymphocytic choriomeningitis virus-infected mice. J. Immunol. 2010, 185, 5549–5560. [Google Scholar] [CrossRef] [PubMed]
- Hoefer, M.M.; Boneberg, E.M.; Grotegut, S.; Kusch, J.; Illges, H. Possible tetramerisation of the proteasome maturation factor Pomp/proteassemblin/hUmp1 and its subcellular localisation. Int. J. Biol. Macromol. 2006, 38, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Salomons, F.A.; Acs, K.; Dantuma, N.P. Illuminating the ubiquitin/proteasome system. Exp. Cell Res. 2010, 316, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Fricke, B.; Heink, S.; Steffen, J.; Kloetzel, P.M.; Kruger, E. The proteasome maturation protein Pomp facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Rep. 2007, 8, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Haas, W.; Crosas, B.; Santamaria, P.G.; Gygi, S.P.; Walz, T.; Finley, D. The heat repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 2005, 12, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.; Bernard Heymann, J.; Kajava, A.V.; Ustrell, V.; Rechsteiner, M.; Steven, A.C. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 2005, 346, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Raveh, D. Proteasome plasticity. FEBS Lett. 2005, 579, 3214–3223. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Groll, M. The 19S cap puzzle: A new jigsaw piece. Structure 2012, 20, 387–388. [Google Scholar] [CrossRef] [PubMed]
- Doherty, K.M.; Pride, L.D.; Lukose, J.; Snydsman, B.E.; Charles, R.; Pramanik, A.; Muller, E.G.; Botstein, D.; Moore, C.W. Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action. G3 2012, 2, 943–959. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.J.; Glanemann, C.; Ramos, P.C.; Dohmen, R.J. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20S proteasomes and promote their maturation. J. Biol. Chem. 2007, 282, 34869–34876. [Google Scholar] [CrossRef] [PubMed]
- Savulescu, A.F.; Glickman, M.H. Proteasome activator 200: The heat is on. Mol Cell. Proteomics 2011. [Google Scholar] [CrossRef]
- Laporte, D.; Lebaudy, A.; Sahin, A.; Pinson, B.; Ceschin, J.; Daignan-Fornier, B.; Sagot, I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 2011, 192, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Bajorek, M.; Glickman, M.H. Keepers at the final gates: Regulatory complexes and gating of the proteasome channel. Cell. Mol. Life Sci. 2004, 61, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Tai, H.C.; Besche, H.; Goldberg, A.L.; Schuman, E.M. Characterization of the brain 26S proteasome and its interacting proteins. Front. Mol. Neurosci. 2010. [Google Scholar] [CrossRef]
- Kaganovich, D.; Kopito, R.; Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008, 454, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Bingol, B.; Schuman, E.M. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature 2006, 441, 1144–1148. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Peth, A.; Goldberg, A.L. Keeping proteasomes under control—A role for phosphorylation in the nucleus. Proc. Natl. Acad. Sci. USA 2011, 108, 18573–18574. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Kato, Y.; Hirano, H. N-myristoylation of the Rpt2 subunit regulates intracellular localization of the yeast 26S proteasome. Biochemistry 2012, 51, 8856–8866. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enenkel, C. Nuclear Transport of Yeast Proteasomes. Biomolecules 2014, 4, 940-955. https://doi.org/10.3390/biom4040940
Enenkel C. Nuclear Transport of Yeast Proteasomes. Biomolecules. 2014; 4(4):940-955. https://doi.org/10.3390/biom4040940
Chicago/Turabian StyleEnenkel, Cordula. 2014. "Nuclear Transport of Yeast Proteasomes" Biomolecules 4, no. 4: 940-955. https://doi.org/10.3390/biom4040940





