Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification
Abstract
:1. tRNA Methylation by the Dnmt2 Family of RNA Methyltransferases
1.1. The Discovery of tRNA Methylation by Dnmt2
1.2. Dnmt2 in the Fission Yeast Schizosaccharomyces pombe: In Vitro Activity
1.3. In Vivo Regulation of Dnmt2 in S. pombe by Nutritional Signals
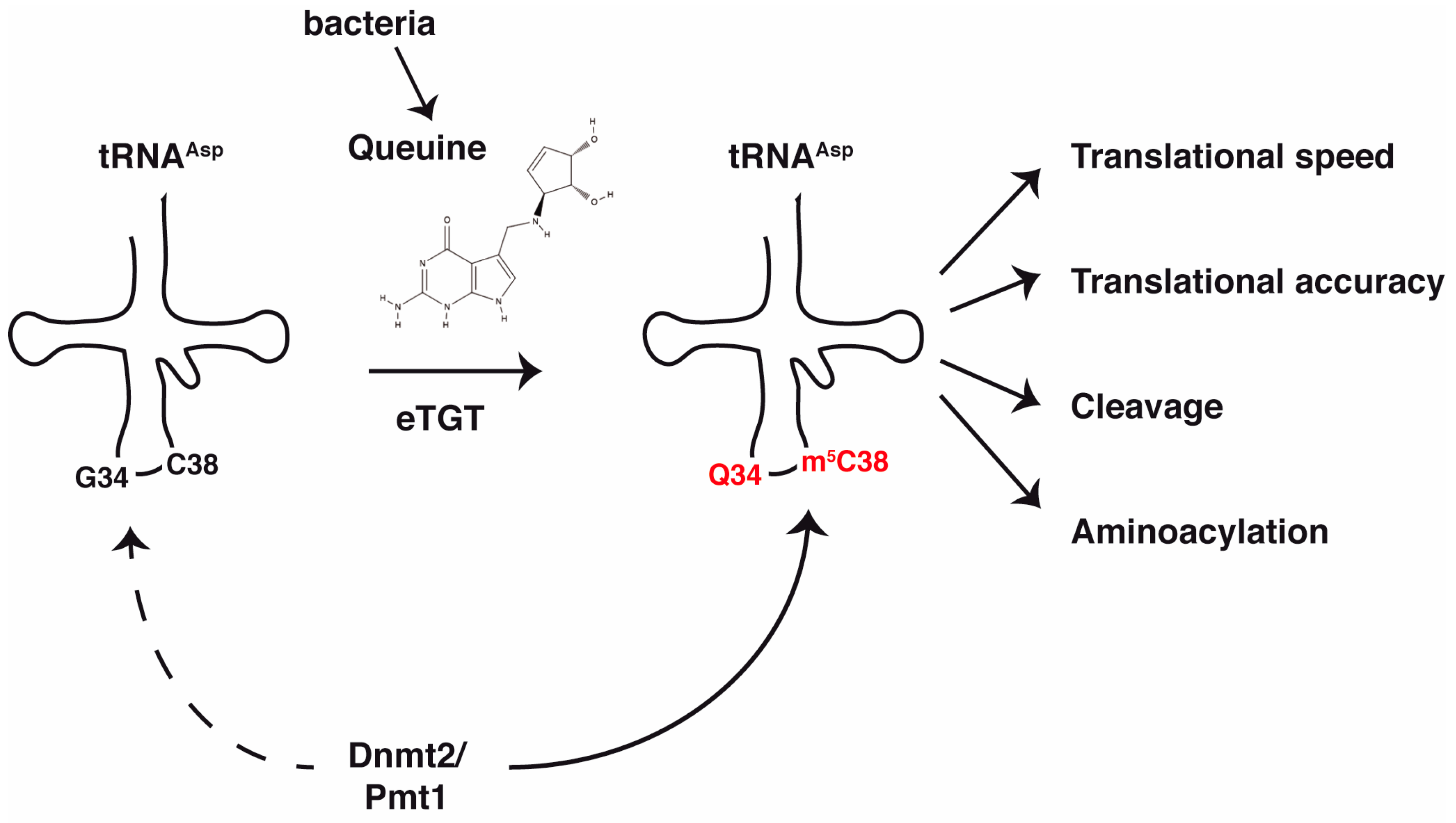
1.4. Stimulation of Dnmt2 Activity by Prior Queuine Incorporation at G34 of tRNAAsp
1.5. Queuine-Independent Stimulation of Dnmt2-Mediated tRNA Methylation Is Mediated by Kinase Signaling
2. Queuosine Modification on tRNAs
2.1. Queuine—Synthesis in Bacteria
2.2. Queuosine Modifications in Eukaryotes
3. Molecular Consequences of Queuosine Modification and Dnmt2-Dependent tRNA Methylation
3.1. Effect of Queuosine on the Dnmt2 Enzyme
3.2. Influence of m5C38 Methylation and Queuosine Modification on Translation
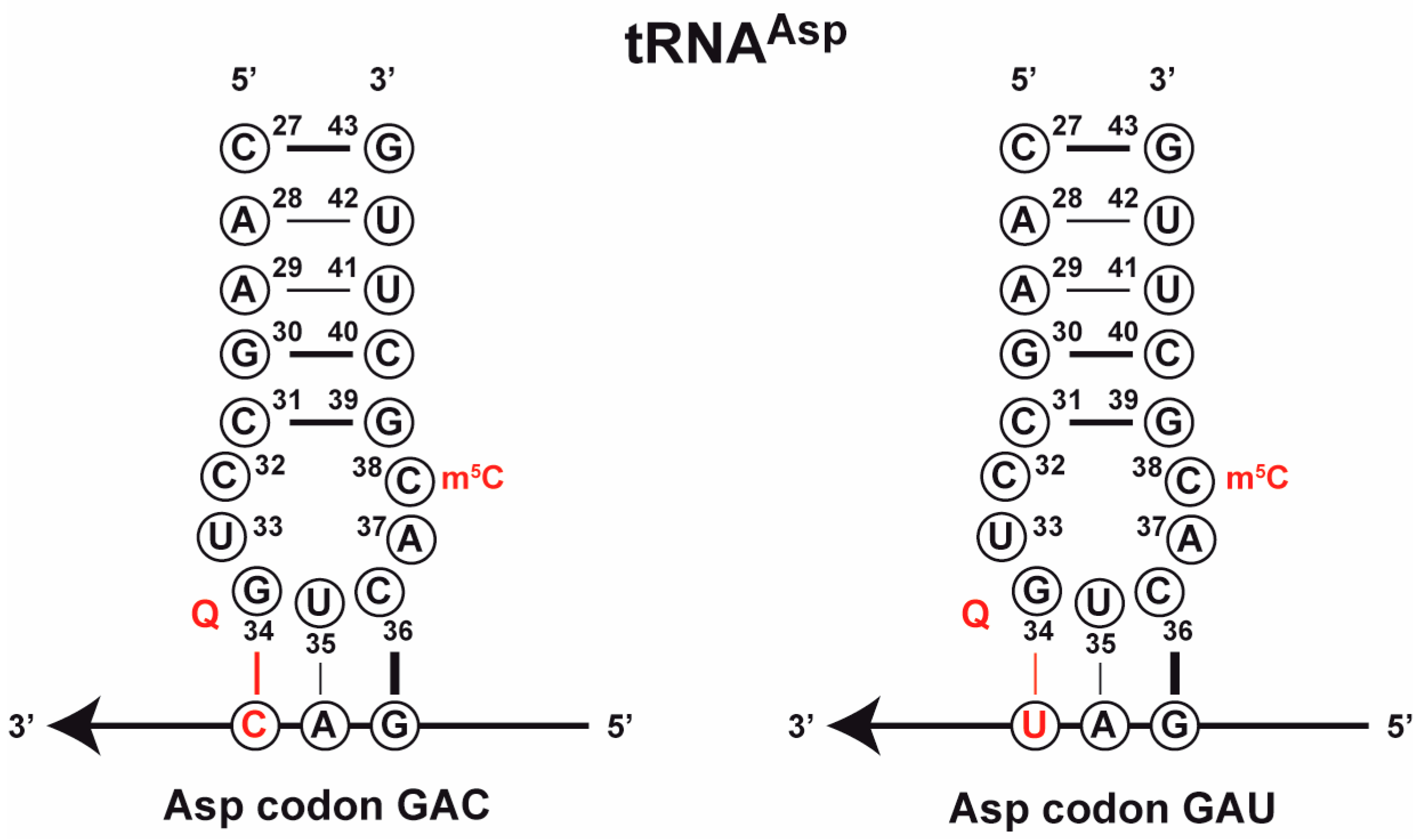
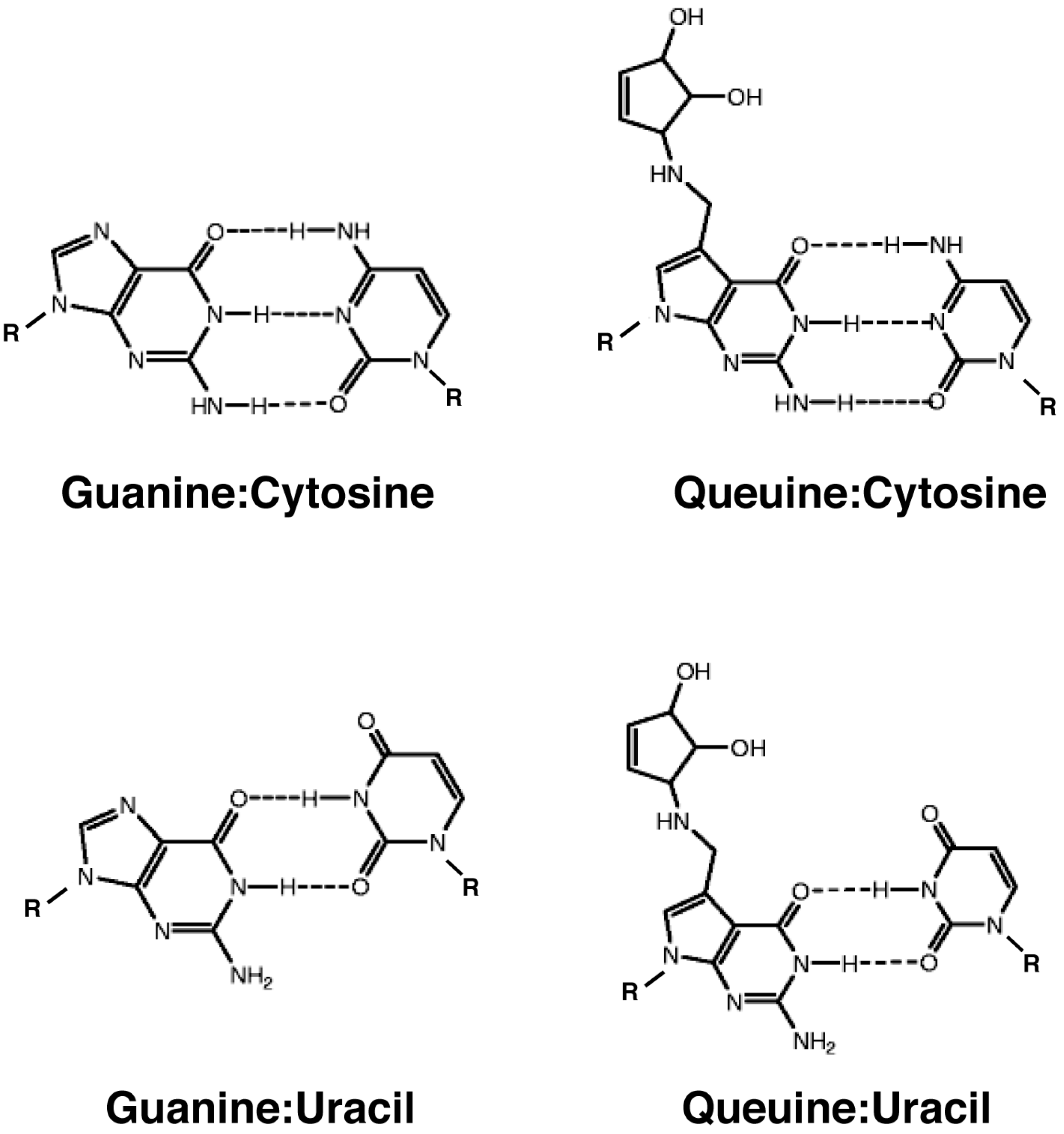
3.2.1. Effect on Codon-Anticodon Base Pairing
3.2.2. In Vivo Effects of Q and Dnmt2 on Translation
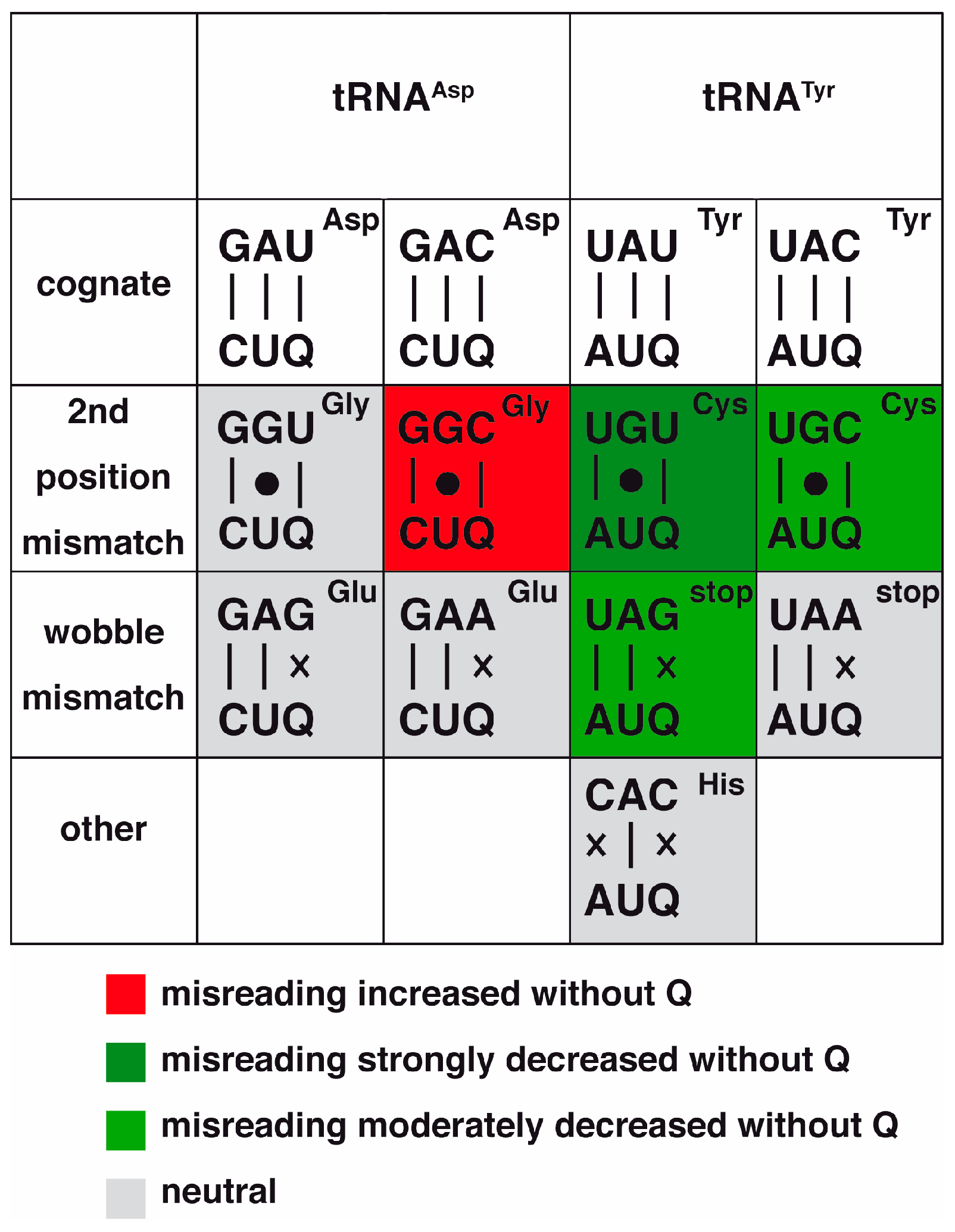
3.2.3. A Role for Queuine and Dnmt2 in Translational Fidelity
3.2.4. Effect Q and m5C38 on the Architecture of the Anticodon Stem-Loop
3.2.5. Effects of Q and m38C on tRNA Aminoacylation
3.2.6. Affinity for the Ribosome
3.2.7. Protection of tRNAs from Endonucleolytic Cleavage by Q and m5C38 Modification
3.3. Evolutionary Conservation of the Dependence of Dnmt2 on Q Modification
4. Organismal Functions of Queuosinylation and C38 Methylation
4.1. Biological Consequences of Dnmt2-Mediated tRNA Methylation
4.1.1. Dnmt2 Mutant Phenotypes in Mice: Roles in Translation and Epigenetic Inheritance
4.1.2. Consequences of Absence of Dnmt2 in Flies: Transposon Silencing, Stress Resistance and Immune Control of Pathogens
4.1.3. Dnmt2 Mutant Phenotypes in Other Organisms
4.2. Organismal Roles for tRNA Queuosinylation
4.3. Combined Phenotypes for the Absence of Dnmt2 and Q
5. Q as a Micronutrient and C38 Modification
5.1. Variations in Q Levels during Development and in Different Organs
5.2. Evolutionary Implications of Q/Dnmt2 Cooperation; Q and Dnmt2 in Natural Selection
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Yoder, J.A.; Bestor, T.H. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 1998, 7, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Hermann, A.; Schmitt, S.; Jeltsch, A. The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 2003, 278, 31717–31721. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Nellen, W.; Lyko, F. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006, 31, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2016, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, R.; Aklujkar, M.; Schafer, M.; Reinhardt, R.; Nickel, O.; Reuter, G.; Lovley, D.R.; Ehrenhofer-Murray, A.; Nellen, W.; Ankri, S.; et al. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res. 2014, 42, 6487–6496. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, T.P.; Jeltsch, A. On the evolutionary origin of eukaryotic DNA methyltransferases and Dnmt2. PLoS ONE 2011, 6, e28104. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.W.; Yu, D.Y.; Pray-Grant, M.G.; Qiu, Q.; Harmon, K.E.; Megee, P.C.; Grant, P.A.; Smith, M.M.; Christman, M.F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 2002, 419, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Antequera, F.; Tamame, M.; Villanueva, J.R.; Santos, T. DNA methylation in the fungi. J. Biol. Chem. 1984, 259, 8033–8036. [Google Scholar] [PubMed]
- Becker, M.; Muller, S.; Nellen, W.; Jurkowski, T.P.; Jeltsch, A.; Ehrenhofer-Murray, A.E. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012, 40, 11648–11658. [Google Scholar] [CrossRef] [PubMed]
- Pinarbasi, E.; Elliott, J.; Hornby, D.P. Activation of a yeast pseudo DNA methyltransferase by deletion of a single amino acid. J. Mol. Biol. 1996, 257, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Hartmann, M.; Schuster, I.; Bender, S.; Thuring, K.L.; Helm, M.; Katze, J.R.; Nellen, W.; Lyko, F.; Ehrenhofer-Murray, A.E. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015, 43, 10952–10962. [Google Scholar] [CrossRef] [PubMed]
- Fergus, C.; Barnes, D.; Alqasem, M.A.; Kelly, V.P. The queuine micronutrient: Charting a course from microbe to man. Nutrients 2015, 7, 2897–2929. [Google Scholar] [CrossRef] [PubMed]
- Katze, J.R.; Basile, B.; McCloskey, J.A. Queuine, a modified base incorporated posttranscriptionally into eukaryotic transfer RNA: Wide distribution in nature. Science 1982, 216, 55–56. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Noguchi, S.; Nishimura, S.; Ohgi, T.; Goto, T.; Crain, P.F.; McCloskey, J.A. Structure determination of a nucleoside Q precursor isolated from E. coli tRNA: 7-(aminomethyl)-7-deazaguanosine. Nucleic Acids Res. 1978, 5, 2289–2296. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Nishimura, Y.; Hirota, Y.; Nishimura, S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 1982, 257, 6544–6550. [Google Scholar] [PubMed]
- Curnow, A.W.; Garcia, G.A. tRNA-guanine transglycosylase from Escherichia coli: Recognition of dimeric, unmodified tRNATyr. Biochimie 1994, 76, 1183–1191. [Google Scholar] [CrossRef]
- Slany, R.K.; Bosl, M.; Kersten, H. Transfer and isomerization of the ribose moiety of AdoMet during the biosynthesis of queuosine tRNAs, a new unique reaction catalyzed by the QueA protein from Escherichia coli. Biochimie 1994, 76, 389–393. [Google Scholar] [CrossRef]
- Miles, Z.D.; McCarty, R.M.; Molnar, G.; Bandarian, V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc. Natl. Acad. Sci. USA 2011, 108, 7368–7372. [Google Scholar] [CrossRef] [PubMed]
- Crain, P.F.; Sethi, S.K.; Katze, J.R.; McCloskey, J.A. Structure of an amniotic fluid component, 7-(4,5-cis-dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanine (queuine), a substrate for tRNA: Guanine transglycosylase. J. Biol. Chem. 1980, 255, 8405–8407. [Google Scholar] [PubMed]
- Farkas, W.R. Effect of diet on the queuosine family of tRNAs of germ-free mice. J. Biol. Chem. 1980, 255, 6832–6835. [Google Scholar] [PubMed]
- Kirtland, G.M.; Morris, T.D.; Moore, P.H.; O’Brian, J.J.; Edmonds, C.G.; McCloskey, J.A.; Katze, J.R. Novel salvage of queuine from queuosine and absence of queuine synthesis in Chlorella pyrenoidosa and Chlamydomonas reinhardtii. J. Bacteriol. 1988, 170, 5633–5641. [Google Scholar] [CrossRef] [PubMed]
- Gaur, R.; Bjork, G.R.; Tuck, S.; Varshney, U. Diet-dependent depletion of queuosine in tRNAs in Caenorhabditis elegans does not lead to a developmental block. J. Biosci. 2007, 32, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Zallot, R.; Brochier-Armanet, C.; Gaston, K.W.; Forouhar, F.; Limbach, P.A.; Hunt, J.F.; de Crecy-Lagard, V. Plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family. ACS Chem. Biol. 2014, 9, 1812–1825. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, K.L.; Seubert, P.H.; Tillman, D.M.; Farkas, W.R.; Katze, J.R. Cloning and characterization of cDNA encoding the rabbit tRNA-guanine transglycosylase 60-kilodalton subunit. Arch. Biochem. Biophys. 1996, 326, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 2014, 42, 7346–7357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Kelly, V.P.; Stachura, S.V.; Garcia, G.A. Characterization of the human tRNA-guanine transglycosylase: Confirmation of the heterodimeric subunit structure. RNA 2010, 16, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Jakobi, S.; Nguyen, T.X.; Debaene, F.; Metz, A.; Sanglier-Cianferani, S.; Reuter, K.; Klebe, G. Hot-spot analysis to dissect the functional protein-protein interface of a tRNA-modifying enzyme. Proteins 2014, 82, 2713–2732. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Brooks, A.F.; Goodenough-Lashua, D.M.; Kittendorf, J.D.; Showalter, H.D.; Garcia, G.A. Evolution of eukaryal tRNA-guanine transglycosylase: Insight gained from the heterocyclic substrate recognition by the wild-type and mutant human and Escherichia coli tRNA-guanine transglycosylases. Nucleic Acids Res. 2011, 39, 2834–2844. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Pais de Barros, J.P.; Keith, G.; Baranowski, W.; Desgres, J. Determination of queuosine derivatives by reverse-phase liquid chromatography for the hypomodification study of Q-bearing tRNAs from various mammal liver cells. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2004, 801, 237–247. [Google Scholar] [CrossRef]
- Dubois, D.Y.; Blaise, M.; Becker, H.D.; Campanacci, V.; Keith, G.; Giege, R.; Cambillau, C.; Lapointe, J.; Kern, D. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp. Proc. Natl. Acad. Sci. USA 2004, 101, 7530–7535. [Google Scholar] [CrossRef] [PubMed]
- Blaise, M.; Olieric, V.; Sauter, C.; Lorber, B.; Roy, B.; Karmakar, S.; Banerjee, R.; Becker, H.D.; Kern, D. Crystal structure of glutamyl-queuosine tRNAAsp synthetase complexed with l-glutamate: Structural elements mediating tRNA-independent activation of glutamate and glutamylation of tRNAAsp anticodon. J. Mol. Biol. 2008, 381, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.; de Crecy-Lagard, V. Biosynthesis and function of tRNA modifications in Archaea. Curr. Opin. Microbiol. 2011, 14, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Du, J.; Yang, H.; Yin, J.; Ding, J.; Zhong, J. Functional and structural characterization of Dnmt2 from Spodoptera frugiperda. J. Mol. Cell Biol. 2013, 5, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.C.; Roth, H.M.; Ankri, S.; Ficner, R. Structure analysis of Entamoeba histolytica Dnmt2 (EhMeth). PLoS ONE 2012, 7, e38728. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.; Yoder, J.A.; Zhang, X.; Zhou, L.; Bestor, T.H.; Cheng, X. Structure of human Dnmt2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001, 29, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Jurkowski, T.P.; Shanmugam, R.; Helm, M.; Jeltsch, A. Mapping the tRNA binding site on the surface of human Dnmt2 methyltransferase. Biochemistry 2012, 51, 4438–4444. [Google Scholar] [CrossRef] [PubMed]
- GtRNAdb, tRNAscan-SE analysis of complete genomes. Available online: http://gtrnadb.ucsc.edu/ (accessed on 2 February 2017).
- Varani, G.; McClain, W.H. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T.; Carmi, A.; Vestsigian, K.; Navon, S.; Dorfan, Y.; Zaborske, J.; Pan, T.; Dahan, O.; Furman, I.; Pilpel, Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 2010, 141, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P. Codon usage bias from tRNA’s point of view: Redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004, 14, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Bailes, E.; Grocock, R.J.; Peden, J.F.; Sockett, R.E. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005, 33, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Ran, W.; Higgs, P.G. The influence of anticodon-codon interactions and modified bases on codon usage bias in bacteria. Mol. Biol. Evol. 2010, 27, 2129–2140. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.B.; Farabaugh, P.J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 2007, 13, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Gilchrist, M.A. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet. 2010, 6, e1001128. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Lyngdoh, R.H. Role of wobble base pair geometry for codon degeneracy: Purine-type bases at the anticodon wobble position. J. Mol. Model. 2012, 18, 3805–3820. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.J.; de Henau, S.; Crothers, D.M. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. USA 1978, 75, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Demeshkina, N.; Jenner, L.; Westhof, E.; Yusupov, M.; Yusupova, G. A new understanding of the decoding principle on the ribosome. Nature 2012, 484, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Rozov, A.; Demeshkina, N.; Westhof, E.; Yusupov, M.; Yusupova, G. Structural insights into the translational infidelity mechanism. Nat. Commun. 2015, 6, 7251. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Suter, B.; Grosjean, H.; Keith, G.; Kubli, E. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J. 1985, 4, 823–827. [Google Scholar] [PubMed]
- Shanmugam, R.; Fierer, J.; Kaiser, S.; Helm, M.; Jurkowski, T.P.; Jeltsch, A. Cytosine methylation of tRNA-Asp by Dnmt2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015, 1, 15010. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Herbst, F.; Alerasool, N.; Bender, S.; Popp, O.; Federico, G.; Reitter, S.; Liebers, R.; Stoecklin, G.; Grone, H.J.; et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015, 34, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Manickam, N.; Nag, N.; Abbasi, A.; Patel, K.; Farabaugh, P.J. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 2014, 20, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Manickam, N.; Joshi, K.; Bhatt, M.J.; Farabaugh, P.J. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res. 2016, 44, 1871–1881. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.C.; Ambrogelly, A.; Crain, P.F.; McCloskey, J.A.; Soll, D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc. Natl. Acad. Sci. USA 2004, 101, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 2016, 44, 8020–8040. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2011, 2, 611–631. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.P.; Vakharia, V.N. The role of queuine in the aminoacylation of mammalian aspartate transfer RNAs. Nucleic Acids Res. 1983, 11, 4257–4272. [Google Scholar] [CrossRef] [PubMed]
- Satwika, D.; Klassen, R.; Meinhardt, F. Anticodon nuclease encoding virus-like elements in yeast. Appl. Microbiol. Biotechnol. 2012, 96, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, G. Anticodon nucleases. Trends Biochem. Sci. 2000, 25, 70–74. [Google Scholar] [CrossRef]
- Frohloff, F.; Fichtner, L.; Jablonowski, D.; Breunig, K.D.; Schaffrath, R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001, 20, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Johansson, M.J.; Bystrom, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Tomita, K.; Ueda, T.; Watanabe, K.; Uozumi, T.; Masaki, H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 1999, 283, 2097–2100. [Google Scholar] [CrossRef] [PubMed]
- Yajima, S.; Inoue, S.; Ogawa, T.; Nonaka, T.; Ohsawa, K.; Masaki, H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res. 2006, 34, 6074–6082. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, J.J.; Kellner, S.M.; Yuan, Y.; Hutinet, G.; Thiaville, P.C.; Jumpathong, W.; Mohapatra, S.; Brochier-Armanet, C.; Letarov, A.V.; Hillebrand, R.; et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, E1452–E1459. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.R.; Bartlett, R.; Nurse, P.; Bird, A.P. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995, 23, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Lyko, F. Solving the Dnmt2 enigma. Chromosoma 2010, 119, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Chidester, S.; Zavala, C.V.; Manos, E.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007, 21, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarian, H.; Wagner, N.; Polo, B.; Baudouy, D.; Kiani, J.; Michiels, J.F.; Cuzin, F.; Rassoulzadegan, M.; Wagner, K.D. Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth. PLoS ONE 2016, 11, e0156953. [Google Scholar] [CrossRef] [PubMed]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiani, J.; Grandjean, V.; Liebers, R.; Tuorto, F.; Ghanbarian, H.; Lyko, F.; Cuzin, F.; Rassoulzadegan, M. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013, 9, e1003498. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Kunert, N.; Marhold, J.; Stanke, J.; Stach, D.; Lyko, F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development 2003, 130, 5083–5090. [Google Scholar] [CrossRef] [PubMed]
- Phalke, S.; Nickel, O.; Walluscheck, D.; Hortig, F.; Onorati, M.C.; Reuter, G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009, 41, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Lyko, F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat. Genet. 2010, 42, 920–921. [Google Scholar] [CrossRef] [PubMed]
- Durdevic, Z.; Hanna, K.; Gold, B.; Pollex, T.; Cherry, S.; Lyko, F.; Schaefer, M. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013, 14, 269–275. [Google Scholar] [CrossRef] [PubMed]
- LePage, D.P.; Jernigan, K.K.; Bordenstein, S.R. The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ 2014, 2, e678. [Google Scholar] [CrossRef] [PubMed]
- Durdevic, Z.; Mobin, M.B.; Hanna, K.; Lyko, F.; Schaefer, M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013, 4, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.J.; Tang, L.Y.; Reddy, M.N.; Shen, C.K. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J. Biol. Chem. 2005, 280, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Yadlapalli, S.; Yamashita, Y.M. Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature 2013, 498, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Arya, D.; Kapoor, S.; Kapoor, M. Physcomitrella patens DNA methyltransferase 2 is required for recovery from salt and osmotic stress. FEBS J. 2016, 283, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Ehrenhofer-Murray, A.E. Humboldt-Universität zu Berlin, Berlin, Germany. Unpublished work. 2016. [Google Scholar]
- Kuhlmann, M.; Borisova, B.E.; Kaller, M.; Larsson, P.; Stach, D.; Na, J.; Eichinger, L.; Lyko, F.; Ambros, V.; Soderbom, F.; et al. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nucleic Acids Res. 2005, 33, 6405–6417. [Google Scholar] [CrossRef] [PubMed]
- Reyniers, J.P.; Pleasants, J.R.; Wostmann, B.S.; Katze, J.R.; Farkas, W.R. Administration of exogenous queuine is essential for the biosynthesis of the queuosine-containing transfer RNAs in the mouse. J. Biol. Chem. 1981, 256, 11591–11594. [Google Scholar] [PubMed]
- Marks, T.; Farkas, W.R. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem. Biophys. Res. Commun. 1997, 230, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Rakovich, T.; Boland, C.; Bernstein, I.; Chikwana, V.M.; Iwata-Reuyl, D.; Kelly, V.P. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J. Biol. Chem. 2011, 286, 19354–19363. [Google Scholar] [CrossRef] [PubMed]
- Ott, G.; Kersten, H.; Nishimura, S. Dictyostelium discoideum: A useful model system to evaluate the function of queuine and of the Q-family of tRNAs. FEBS Lett. 1982, 146, 311–314. [Google Scholar] [CrossRef]
- Siard, T.J.; Jacobson, K.B.; Farkas, W.R. Queuine metabolism and cadmium toxicity in Drosophila melanogaster. BioFactors 1991, 3, 41–47. [Google Scholar] [PubMed]
- Durand, J.M.; Okada, N.; Tobe, T.; Watarai, M.; Fukuda, I.; Suzuki, T.; Nakata, N.; Komatsu, K.; Yoshikawa, M.; Sasakawa, C. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J. Bacteriol. 1994, 176, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Durand, J.M.; Dagberg, B.; Uhlin, B.E.; Bjork, G.R. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: The expression of the virF gene. Mol. Microbiol. 2000, 35, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Hurt, J.K.; Olgen, S.; Garcia, G.A. Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic Acids Res. 2007, 35, 4905–4913. [Google Scholar] [CrossRef] [PubMed]
- Stengl, B.; Meyer, E.A.; Heine, A.; Brenk, R.; Diederich, F.; Klebe, G. Crystal structures of tRNA-guanine transglycosylase (Tgt) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding. J. Mol. Biol. 2007, 370, 492–511. [Google Scholar] [CrossRef] [PubMed]
- Varghese, S.; Cotter, M.; Chevot, F.; Fergus, C.; Cunningham, C.; Mills, K.H.; Connon, S.J.; Southern, J.M.; Kelly, V.P. In vivo modification of tRNA with an artificial nucleobase leads to full disease remission in an animal model of multiple sclerosis. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Vinayak, M.; Pathak, C. Queuosine modification of tRNA: Its divergent role in cellular machinery. Biosci. Rep. 2009, 30, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.B.; Farkas, W.R.; Katze, J.R. Presence of queuine in Drosophila melanogaster: Correlation of free pool with queuosine content of tRNA and effect of mutations in pteridine metabolism. Nucleic Acids Res. 1981, 9, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Zaborske, J.M.; DuMont, V.L.; Wallace, E.W.; Pan, T.; Aquadro, C.F.; Drummond, D.A. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol. 2014, 12, e1002015. [Google Scholar] [CrossRef] [PubMed]
- White, B.N.; Tener, G.M. Activity of a transfer RNA modifying enzyme during the development of Drosophila and its relationship to the su(s) locus. J. Mol. Biol. 1973, 74, 635–651. [Google Scholar] [CrossRef]
- Hiraoka, Y.; Kawamata, K.; Haraguchi, T.; Chikashige, Y. Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells 2009, 14, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Harada, F.; Nishimura, S. Possible anticodon sequences of tRNAHis, tRNAAsm, and tRNAAsp from Escherichia coli B. Universal presence of nucleoside Q in the first postion of the anticondons of these transfer ribonucleic acids. Biochemistry 1972, 11, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yi, C.; Yu, Y.T. Transcriptome-wide dynamics of RNA pseudouridylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 581–585. [Google Scholar] [CrossRef] [PubMed]
| Organism | Dnmt2 Homolog | Dnmt2 Substrate(s) | Presence of TGT |
|---|---|---|---|
| Mus musculus | Dnmt2 | tRNAAsp, tRNAGly, tRNAVal | present |
| Drosophila melanogaster | Dnmt2 | tRNAAsp, tRNAGly, tRNAVal | present |
| Caenorhabditis elegans | none | - | present |
| Dictyostelium discoideum | DnmA | tRNAAsp (in vivo and in vitro), tRNAGlu (only in vitro), tRNAGly (only in vitro) | present |
| Schizosaccharomyces pombe | Dnmt2/Pmt1 | tRNAAsp (depends on Q or Pmt1 overexpression), tRNAGlu (weak, only upon Pmt1 overexpression) | present |
| Saccharomyces cerevisiae | none | - | none |
| Geobacter sulfurreducens | Dnmt2 | tRNAGlu | present |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehrenhofer-Murray, A.E. Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification. Biomolecules 2017, 7, 14. https://doi.org/10.3390/biom7010014
Ehrenhofer-Murray AE. Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification. Biomolecules. 2017; 7(1):14. https://doi.org/10.3390/biom7010014
Chicago/Turabian StyleEhrenhofer-Murray, Ann E. 2017. "Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification" Biomolecules 7, no. 1: 14. https://doi.org/10.3390/biom7010014





