1. Introduction
Sodium-potassium adenosine triphosphatase (Na,K-ATPase) is an enzyme that transports Na
+ and K
+ through the plasma membrane against the electrochemical gradient. The transport of cations is coupled with hydrolysis of adenosine triphosphate (ATP) to adenosine diphosphate (ADP) and inorganic phosphate (P
i ), which supplies the needed energy. The enzyme is composed of two subunits (catalytic α-subunit and regulatory β-subunit) having molecular masses of 110 and 65 kDa, respectively. Four isoforms of the Na,K-ATPase α-subunit and three isoforms of β-subunit are now known [
1]. The isoforms of the two subunits can be combined in different ways creating various heterodimers. The α1β1-isoenzyme is a ‘housekeeping’ enzyme located almost in all animal tissues. The α1β1 enzyme is the only isoenzyme found in tissues providing sodium excretion (mammalian kidney, salt glands of birds). The distribution of other isoforms is tissue-specific.
The polypeptide chain of the Na,K-ATPase α-subunit crosses the membrane 10 times, and its transmembrane segments form a channel with gates for ion transport [
2]. Cytoplasmic part of the polypeptide chain is assembled in three cytoplasmic domains. Two of these (nucleotide binding and phosphorylated domains) participate in the formation of the enzyme active site. The Na,K-ATPase α-subunit is required for transport of the enzyme to the membrane, and it affects K
+ transport [
3].
The α1-subunit contains 23 cysteine residues, 15 of them facing the cytosol. There are no disulfide bonds between cysteine residues of the α-subunit [
4,
5]. Changing cysteine residues to alanine in the α-subunit is not critical for enzyme activity [
6,
7], so the role of the cysteines is not clear. However, Na,K-ATPase is extremely sensitive to intracellular redox status [
4,
8,
9,
10,
11,
12,
13]. This suggests that redox modification of cysteines may play an important role in the regulation of Na,K-ATPase.
We found earlier that Na,K-ATPase from various tissues (rabbit kidney, rat heart, duck salt glands) has a significantly
S-glutathionylated α1-subunit [
8,
9,
10]. The level of the α1-subunit glutathionylation depends on cellular redox status and increases under hypoxia [
8,
10]. In the Na,K-ATPase α1-subunit from duck salt glands, up to 12 of the 15 cysteine residues facing the cytosol can be
S-glutathionylated [
8]. Treatment of Na,K-ATPase from this tissue with oxidized glutathione in vitro resulted in the glutathionylation of Cys-244, -454, -458, and -459 of the α1-subunit, leading to complete inhibition of the enzyme activity. After glutathione binding to these cysteines, the enzyme was unable to interact with adenine nucleotides [
8]; apparently glutathione bound to three cysteine residues (Cys-454, -458, and -459) near the ATP-binding site can prevent ATP binding [
8]. Because adenine nucleotides can prevent the enzyme glutathionylation and ATP is most effective in this respect [
8,
9], depletion of ATP will induce glutathionylation. It was shown that the only free SH-group of β-subunit could bind glutathione, which decreases the enzyme activity by approximately 20% [
14]. Thus, glutathionylation of some cysteines of Na,K-ATPase subunits is involved in the regulation of its activity.
In addition to four regulatory cysteine residues of Na,K-ATPase α1-subunit that are glutathionylated after treatment with oxidized glutathione, many other cysteines are in the glutathionylated form in purified enzyme. The functional significance of their
S-glutathionylation remains unclear. We suggested that glutathionylation of some cysteines of the Na,K-ATPase α-subunit takes place cotranslationally and is important for protein folding [
5]. It was shown earlier that hypoxic rat heart cysteine residues of Na,K-ATPase can be also
S-nitrosylated, and that in hypoxic rat heart there is cross talk between
S-nitrosylation and
S-glutathionylation of the Na,K-ATPase α1-subunit [
15]. Oxidation of SH-groups to -SOH in proteins can proceed to their glutathionylation or nitrosylation, but irreversibly oxidized cysteines (to cys-SO
2H and cys-SO
3H) are now considered as markers for protein degradation [
4].
To better understand the role of various redox modifications of cysteine residues of the Na,K-ATPase α-subunit in its function, we have studied effects of different reducing agents and the enzymatic reducing system glutaredoxin/glutathione reductase on the modified cysteine residues of Na,K-ATPase and on the activity of the enzyme.
2. Results
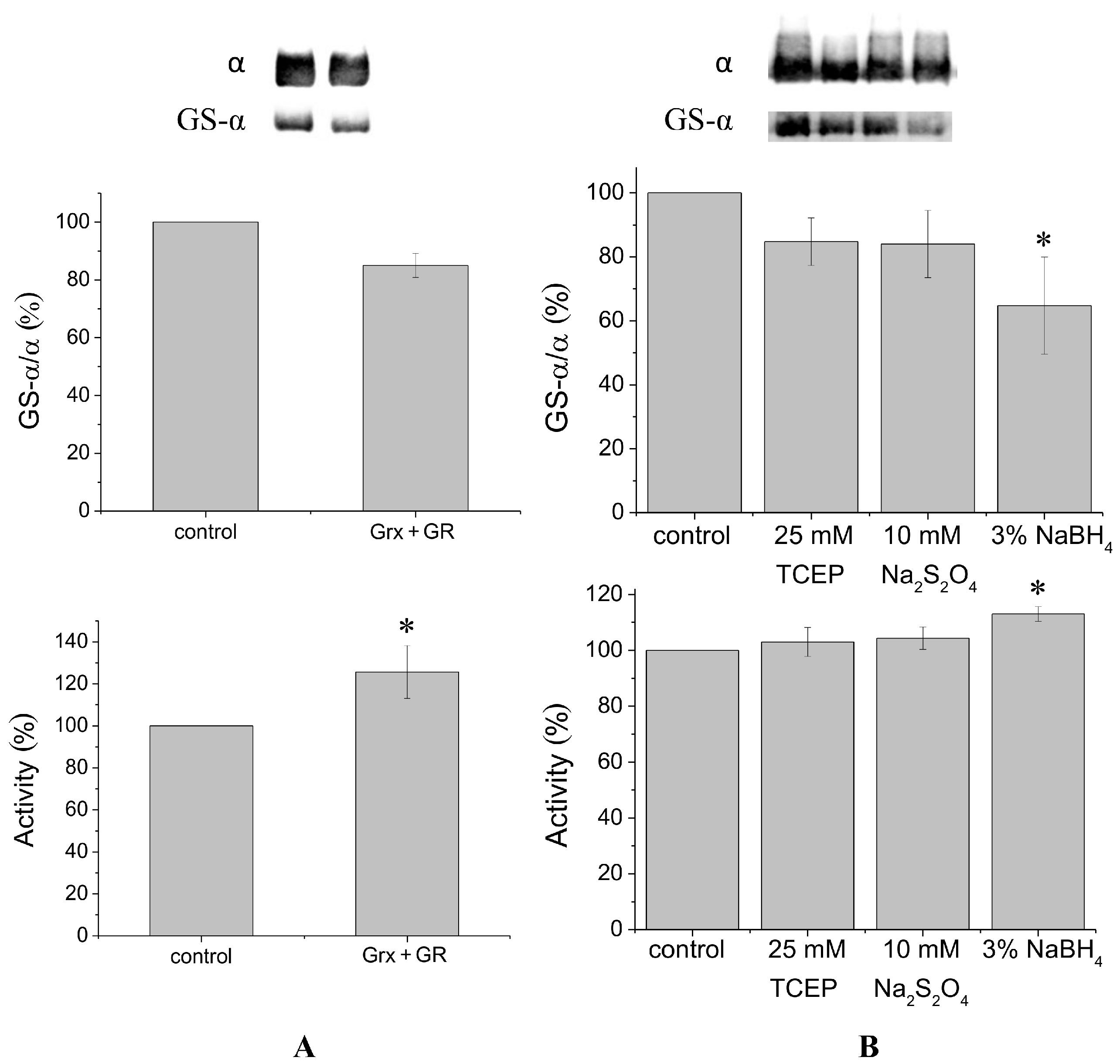
It is known that glutaredoxin combined with glutathione reductase in the presence of their substrates glutathione and nicotinamide adenine dinucleotide phosphate (NADPH) can catalyze protein deglutathionylation [
4,
8]. Incubation of duck salt gland Na,K-ATPase in vitro with glutaredoxin/glutathione reductase for 30–60 min results in a small decrease in the level of Na,K-ATPase α1-subunit glutathionylation detected using antibodies against glutathione bound to proteins (
Figure 1A,
Supplementary Figure S2). Although deglutathionylation of the α1-subunit using glutaredoxin/glutathione reductase does not exceed ~20%, it increases Na,K-ATPase activity by ~30% (
Figure 1A).
To deglutathionylate natively glutathionylated Na,K-ATPase α1-subunit, we also used chemical reducing agents tris(2-carboxyethyl)-phosphine (TCEP), sodium dithionite, and sodium borohydride, that have redox potentials of −0.29, −0.416, and −1.24 V, respectively. We previously found concentrations of these reagents that provide the maximal effect (data not shown). Treatment of Na,K-ATPase by TCEP, sodium dithionite, or sodium borohydride for 30 min at 37 °C decreased glutathionylation level of the α1-subunit by 15%–35% (
Figure 1B,
Supplementary Figure S3). Sodium borohydride, having the lowest redox potential, provided the maximal effect. Dithiothreitol and β-mercaptoethanol, which have redox potentials comparable with TCEP (−0.33 and −0.26 V, respectively) decreased the level of glutathionylation almost as TCEP (by ~12%, see
Supplementary Figure S6). In contrast to enzymatic deglutathionylation, treatment of Na,K-ATPase with reducing agents was accompanied by a slight increase in Na,K-ATPase activity (
Figure 1B). The maximal increase (less than 20%) was observed after treating the enzyme with sodium borohydride (
Figure 1B).
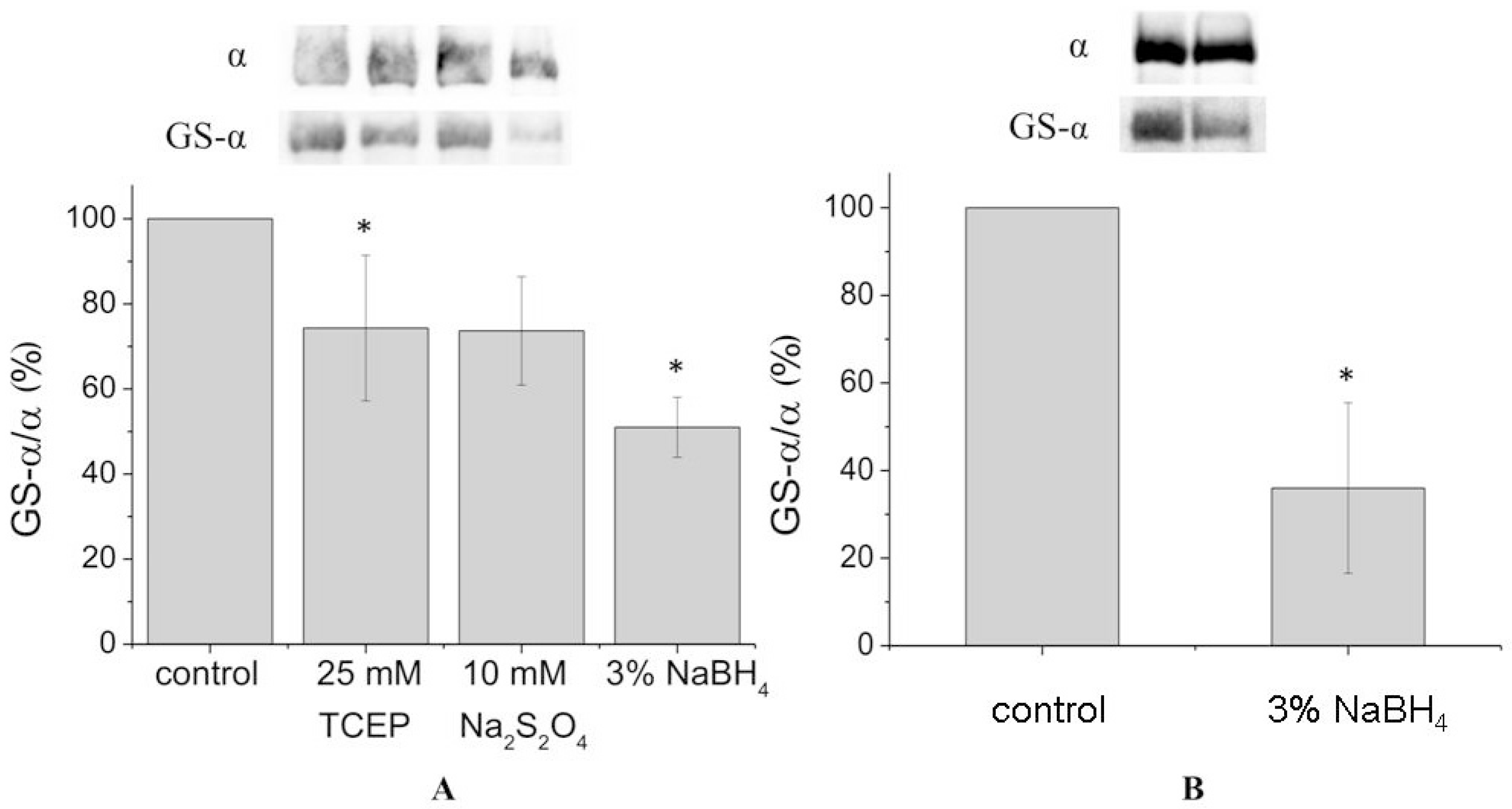
In order to achieve complete deglutathionylation of the α1-subunit, we treated the enzyme with chemical reducing agents under denaturing conditions (in the presence of 8% sodium dodecyl sulfate (SDS) and 8 M urea at 37 °C). However, even under these conditions deglutathionylation of the α1-subunit was partial, not exceeding 50% (
Figure 2A,
Supplementary Figures S4 and S7). For the microsomal fraction, deglutathionylation of the α1-subunit reached 65% (
Figure 2B,
Supplementary Figure S5).
To check for the presence of glutathione bound to the α1-subunit after its treatment with reducing agents, we performed mass spectrometry analysis of enzyme preparation that was incubated with or without sodium borohydride under denaturing conditions (
Table 1 and
Supplementary Tables S1 and S2, sequence coverage was 69.0% and 69.9% correspondingly).
Supplementary Tables contain information about tryptic peptides of α1-subunit with different redox modifications of cysteine residues in the control sample (
Supplement Table S1) and sample treated by sodium borohydride (
Supplement Table S2) under denaturing conditions.
Table 1 shows the numbers of different tryptic peptides (including peptides with sequences having different numbers of trypsin miscleavage sites) with a determined redox modification of the given cysteine residue under control conditions (Control) and after treatment with sodium borohydride (Reduced). Firstly, we should note that patterns of tryptic cleavage of the α1-subunit polypeptide chain before and after enzyme treatment by sodium borohydride were different. In the control preparation, we found 28 different tryptic fragments of the α1-subunit polypeptide chain containing 13 cysteine residues (
Supplementary Table S1). After the reduction of cysteine residues of Na,K-ATPase under denaturing conditions, we ascertained 26 tryptic fragments of the α-subunit containing 13 cysteine residues (
Supplementary Table S2). After the reduction of the denatured protein, we found only five peptides containing
S-glutathionylated cysteine residues (Cys-454, -458, -459, -513, and -658).
Three of the cysteine residues of the α1-subunit of Na,K-ATPase (Cys-140, -206, and -351) after reduction of the denatured enzyme lost their bound glutathiones. We also observed a decrease in the number of peptides containing glutathionylated Cys-454, -458, and -459 after the reduction of the denatured protein.
Cys-423 was found in tryptic fragments of native enzyme only in the unmodified form, which is consistent with our results obtained earlier [
8]. However, we did not observe peptides with modified Cys-369 although earlier we detected a low intensity peak for the peptide with glutathionylated Cys-369 [
8]. Eight cysteine residues of the α1-subunit (Cys-140, -206, -351, -454, -458, -459, -513, and -658) were
S-glutathionylated. There were no peptides with glutathionylated Cys-244 and Cys-700 (
Table 1), which we have detected in other preparations where denaturing conditions did not apply [
8]. However, in the analyzed preparation of Na,K-ATPase, these cysteine residues were oxidized, which could prevent their glutathionylation (
Table 1). We also detected a peptide with glutathionylated Cys-140, which was not revealed earlier [
8]. After reduction under denaturing conditions Cys-140, -206, and -351 lost their bound glutathione. We also observed a decrease in the number of peptides containing glutathionylated Cys-454, -458, and -459 (
Table 1).
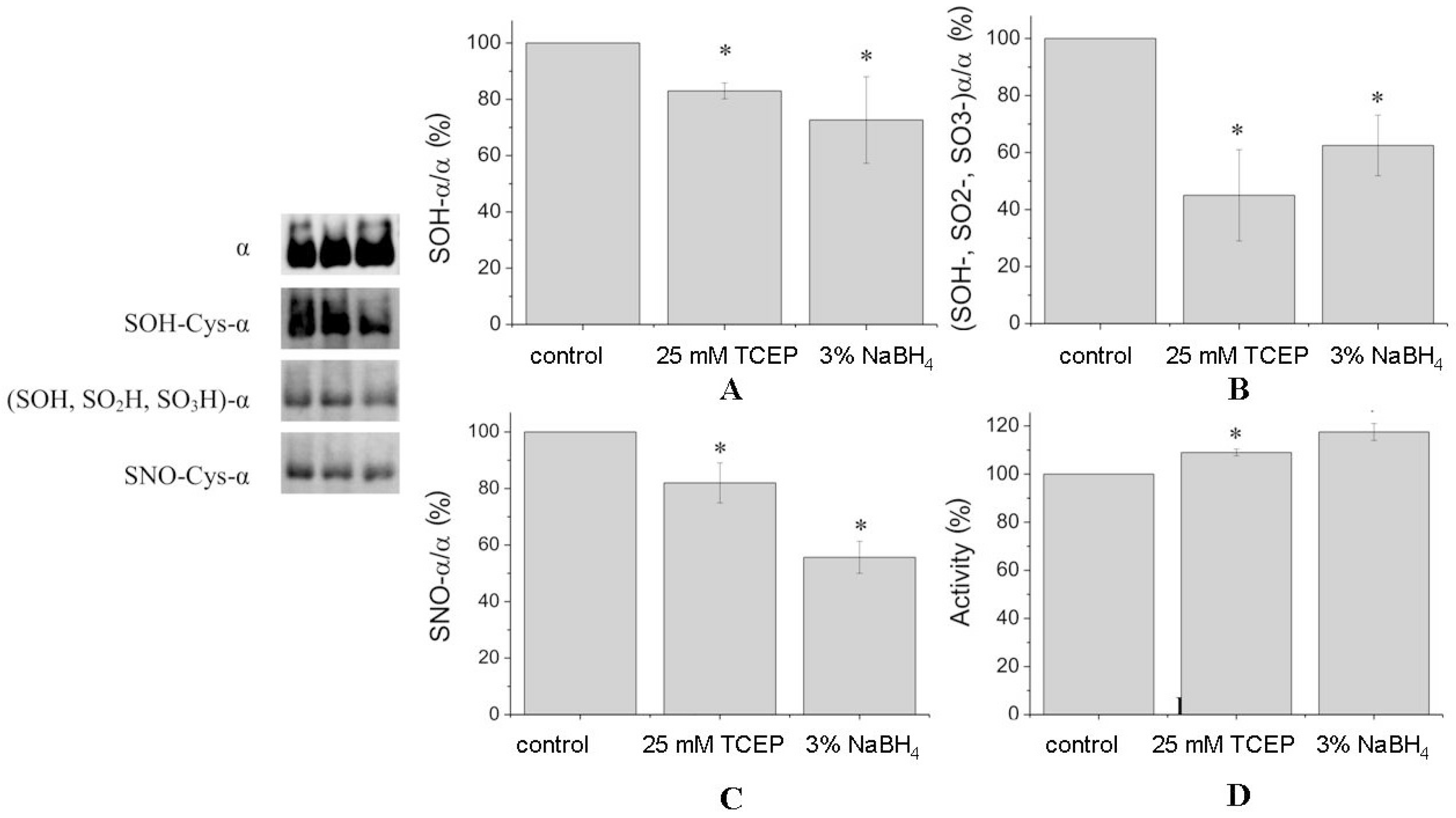
Four cysteine residues of the α1-subunit (Cys-351, -454, -458, -459) were found in nitrosylated forms (
Table 1). Nitrosylated Cys-700 was revealed only in the preparation after reduction. This may be due to the fact that the pattern of tryptic cleavage after reduction was changed and we did not detect a peptide corresponding to nitrosylated Cys-700 in the reduced preparation. After treatment of denaturated protein by reducing agents, the number of nitrosylation of cysteine residues has not changed but the total number of nitrosylated peptides decreased. Treatment of undenatured Na,K-ATPase by TCEP and NaBH
4 decreased the amount of Cys-SNO by 20% to 57%, correspondingly (
Figure 3C).
Some cysteines were oxidized to Cys-SOH and Cys-SO
2H (
Table 1). TCEP and sodium borohydride decreased the amount of Cys-SOH in α1-subunit by 20% to 30% (
Figure 3A). However, the level of all oxidized cysteines (Cys-SOH, Cys-SO
2H, Cys-SO
3H) of the α1-subunit was decreased by TCEP and sodium borohydride by ~60% and ~40%, respectively (
Figure 3B).
It can be seen that many cysteines of the α1-subunit had several different redox modifications. Cys-351 that had all types of redox modifications was found after the reduction only in the unmodified and nitrosylated forms. Peptides including Cys-456, -458, and -459 before and after treatment of protein with reducing agents were found in unmodified forms and with all types of cysteine residue modifications, although the total number of modified peptides was decreased (
Table 1).
Effects of reducing agents on all the modifications of cysteine residues studied using antibodies against specific modifications of cysteines were confirmed by mass spectrometry. The data presented in
Table 1 demonstrate that the total number of peptides containing all types of redox modifications of cysteines of α1-subunit dropped about two-fold after the reduction of denatured Na,K-ATPase by sodium borohydride.
3. Discussion
The Na,K-ATPase α1-subunit from different sources has 23 cysteine residues [
4]. Individual replacement of cysteines by alanine or by serine in the α1-subunit resulted in some cases in decrease of the enzyme turnover by 2–4-fold [
6]. However, replacement of Cys-242 (Cys-244 in duck α1-subunit) led to the cessation of the enzyme expression. Molecular activity of the cysteine-less (cysteine residues in the α-subunit were substituted with alanines or serines) mutants of Na,K-ATPase and the K
m values for activation by ATP were similar to the values characteristic for the wild-type enzyme [
7]. However, a number of parameters for cysteine-less enzyme were different: 50% reduction of K
m for Na
+, and 2-fold increase in value for K
m for K
+ were observed. Moreover, the stability of cysteine-less enzyme was decreased significantly. The cysteine-less enzyme has higher content in endoplasmic reticulum and Golgi apparatus in comparison with the wild-type protein, it has lower stability [
7]. Thus, cysteines are not critical for function, but some of them appear to be necessary for α1-subunit folding, trafficking, assembly, and stability [
7]. Despite the replacement of cysteine residues, enzyme activity does not change [
6,
7]; however, modification of regulatory cysteine residues by glutathione can result in complete enzyme inhibition [
8]. We suggest that cysteines are important for redox-sensitive regulation of Na,K-ATPase. Oxidative modifications of Na,K-ATPase thiols may also be involved in oxygen-inducible regulation of enzyme functions [
4,
8].
We showed that in addition to regulatory glutathionylation of the α1-subunit (Cys-454, -458, -459, -244) (
Supplementary Figure S8), basal glutathionylation of the Na,K-ATPase that cannot be removed in the native protein also exists [
5]. Unresolved density in isolated cavities in X-ray structure of pig Na,K-ATPase can be occupied by molecules of glutathione associated with the pairs of cysteine residues 204–242, 367–698, and 452–456 (206–244, 369–700, and 454–458 in the duck α1-subunit, which are located in actuator (A), nucleotide binding (N), and phosphorylated (P) domains respectively (
Supplementary Figure S8) [
4,
5]. According to our previous data [
5], cysteine residues 204, 452, and 698 are preferable for basal glutathionylation. It should be noted that each of the domains (A, N, and P) contains basal glutathionylated residue (
Supplementary Figure S8). We suggested that basal glutathionylation can prevent the formation of disulphide bridges between the neighboring cysteine residues during folding of α-subunit [
5]. Formation of disulfide bridges can increase structure rigidity and prevents conformational lability of the molecule, needed for moving Na,K-ATPase domains in catalytic cycles. We assumed that this basal glutathionylation occurs cotranslationally and ‘remembers’ the cellular redox state that existed during protein biosynthesis.
According to our data (
Figure 1) application of coupled enzyme system glutaredoxin/glutathione reductase unlike chemical reducing agents leads to significant increase of enzyme activity. At the same time the enzyme system glutaredoxin/glutathione reductase deglutathionylates native Na,K-ATPase in a lesser degree than chemical reducing agents, so the enzyme system deglutathionylates only some regulatory thiol groups [
8].
We have not been able to fully remove bound glutathiones from the denatured purified α-subunit even by applying borohydride. However, we completely removed glutathionylation in the denatured murine α1-subunit previously [
5]. This can be explained by differences in the microenvironment of cysteines in α1-subunits from duck and mouse which can change their susceptibility to glutathionylation. The nearest-neighbor residues to cysteines in the α1-subunit sequence affect the pK
a of Cys thiol group, changing its reactivity [
4]. Another example of an extra stable disulfide bond, which was not disrupted even at 60 °C in the presence of 10 mM dithiothreitol appears in the double cysteine mutant of subtilisin between Cys-22 and Cys-87 [
16].
Taking into account all obtained data, we can suggest the following concerning the function of glutathionylation of cysteine residues of Na,K-ATPase α1-subunit. Short time oxidative stress and short time hypoxia that lead to the increase of GSSG (oxidazed glutathione, hexapeptide) concentrations results in the increase of glutathionylation of regulatory cysteines in the α-subunit (located in the N or A domains,
Figure S8) that inhibits the enzyme and prevents ATP exhaustion in the cells [
8]. At the same time, prolonged oxidative stress or prolonged hypoxia lead to the glutathionylation of some cysteines that proceeds during protein folding. This increase of basic glutathionylation of Na,K-АТPase α1-subunit that cannot be removed from native protein [
5], because the modified residues (located in the A, N, and P domains,
Supplementary Figure S8) are inaccessible for reduction agents. We suggest that increase of basic glutathionylation results in the change of protein properties that, in turn, may be important for the adaptation to prolonged oxidative stress. At the same time, the glutathionylation of non-regulatory (but accessible for reducing agents) cysteine residues will protect the enzyme from irreversible oxidation. We have found that, in Na,K-АТPase purified from duck salt glands, there is some glutathionylated cysteine residues reduction of which using enzyme system results in the increase of enzyme activity. We have also found some cysteine residues whose deglutathionylation does not affect enzyme activity. Additionally, we have found some basically glutathionylated cysteine residues that cannot be deglutathionylated under different conditions. Only Cys-423 buried in N domain (
Supplementary Figure S8) both previously [
8] and now (
Table 1) was found in the unmodified form. Therefore, we can place cysteine residues of Na,K-ATPase α1-subunit into three functional groups: regulatory cysteines (responsible for the enzyme activity, their glutathionylation leads to enzyme inhibition), basically glutathionylated residues that are important for protein folding and prolonged adaptation, and normal cysteine residue glutathionylation which does not affect the activity, but protects thiol groups against irreversible oxidation.
Earlier, it was hypothesized that nitrosylation of cysteine residues was important for maintaining high activity of Na,K-ATPase at physiological oxygen levels [
11,
12,
13], and that cross talk existed between glutathionylation and nitrosylation of the Na,K-ATPase catalytic subunit. This was demonstrated previously [
15]. If nitric oxide synthases activity is decreased during hypoxia, the level of α1-subunit nitrosylation is also decreased, whereas the level of glutathionylation is increased [
8,
10]. Both modifications protect cysteine residues from irreversible oxidation to sulfinic and sulfonic acids. However, glutathionylation, in contrast to nitrosylation, leads to enzyme inhibition, which can be reversed when the cell returns to the normal redox state using some enzyme coupled systems such as glutaredoxin/glutathione reductase.
Indeed, we observed not only glutathionylation but also nitrosylation of a number of cysteine residues in the α1-subunit of Na,K-ATPase from duck salt glands. The most nitrosylated cysteine residues are the same cysteine residues that can be glutathionylated, which is consistent the with cross-talk hypothesis. Treatment of Na,K-ATPase with reducing agents decreases the degree of nitrosylation of cysteine residues but does not eliminate nitrosylation completely, as well as glutathionylation. Thiol groups of cysteine residues that are not protected by nitrosylation or by glutathionylation seem to be oxidized to sulfenic, sulfinic, and sulfonic acids.
It should be noted that Cys-454, -458, and -459 (two of them are located in a cavity) lying near the ATP-binding site (
Supplementary Figure S8) exist in nitrosylated, glutathionylated, and oxidized forms before and after reduction, and these modifications cannot be elicited by reducing agents. This supports our hypothesis that some of these cysteines are involved in the regulation of Na,K-ATPase activity [
8] but the others are not involved in the enzyme activity regulation since they are located in isolated cavities [
5]. Being glutathionylated during hypoxia, they inhibit the enzyme, but after in vivo deglutathionylation by the glutaredoxin/glutathione reductase enzyme system under normal conditions, they are nitrosylated to prevent irreversible deep (to -SO
2H and -SO
3H) oxidation.
Cysteines of the Na,K-ATPase α1-subunit can be oxidized to different levels. Irreversible oxidation of cysteines of the protein to Cys-SO
3H and Cys-SO
2H was proposed as a marker for protein degradation by the cellular machinery [
17] whereas oxidation to Cys-SOH is considered now as a posttranslational modification that can be important for enzyme regulation [
18]. Treatment of Na,K-ATPase with reducing agents decreases the number of Cys-SOH and Cys-SO
2H (
Table 1)
. This correlates with data of Western blotting, where the amount of all oxidized forms of cysteines is decreased more significantly than the amount of Cys-SOH (
Figure 3).
Thus, using two methods, we have found that a number of cysteine residues of Na,K-ATPase purified from duck salt glands are glutathionylated, nitrosylated, and oxidized to a different degree. Reducing agents decreased the level of all these modifications (but not completely) leading to a slight increase in the enzyme activity. The natural enzyme system glutaredoxin/glutathione reductase that decreased glutathionylation to a lesser extent than the reducing agents produced a greater increase in enzyme activity. This suggests that the enzyme deglutathionylation system specifically affects glutathionylation of the regulatory cysteine residues of Na,K-ATPase α-subunit.