Common Chemical Inductors of Replication Stress: Focus on Cell‐Based Studies
Abstract
:1. Introduction
2. Compounds
2.1. Cisplatin
2.1.1. Mechanism of DNA Damage Induction
2.1.2. Other Effects
2.1.3. Solubility
2.1.4. Medical Use
2.1.5. Summary
2.2. Aphidicolin
2.2.1. Mechanism of DNA Damage Induction
2.2.2. Other Effects
2.2.3. Solubility
2.2.4. Medical Use
2.2.5. Summary
2.3. Hydroxyurea
2.3.1. Mechanism of DNA Damage Induction
2.3.2. Other Effects
2.3.3. Solubility
2.3.4. Medical Use
2.3.5. Summary
2.4. Camptothecin
2.4.1. Mechanism of DNA damage induction
2.4.2. Solubility
2.4.3. Medical Use
2.4.4. Summary
2.5. Etoposide
2.5.1. Mechanism of DNA Damage Induction
2.5.2. Other Effects
2.5.3. Solubility
2.5.4. Medical Use
2.5.5. Summary
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zeman, M.K.; Cimprich, K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Burhans, W.C.; Weinberger, M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007, 35, 7545–7556. [Google Scholar] [CrossRef] [PubMed]
- Huh, M.S.; Ivanochko, D.; Hashem, L.E.; Curtin, M.; Delorme, M.; Goodall, E.; Yan, K.; Picketts, D.J. Stalled replication forks within heterochromatin require ATRX for protection. Cell Death Dis. 2016, 7, e2220. [Google Scholar] [CrossRef] [PubMed]
- Gelot, C.; Magdalou, I.; Lopez, B.S. Replication stress in Mammalian cells and its consequences for mitosis. Genes 2015, 6, 267–298. [Google Scholar] [CrossRef] [PubMed]
- Krasilnikova, M.M.; Mirkin, S.M. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004, 24, 2286–2295. [Google Scholar] [CrossRef] [PubMed]
- Neelsen, K.J.; Zanini, I.M.Y.; Mijic, S.; Herrador, R.; Zellweger, R.; Ray Chaudhuri, A.; Creavin, K.D.; Blow, J.J.; Lopes, M. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013, 27, 2537–2542. [Google Scholar] [CrossRef] [PubMed]
- Porter, A.C. Preventing DNA over-replication: A Cdk perspective. Cell Div. 2008, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.-C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Alberts, B.M. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 1995, 267, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Rezaei, N.; Liontos, M.; Karakaidos, P.; Kletsas, D.; Issaeva, N.; Vassiliou, L.-V.F.; Kolettas, E.; Niforou, K.; Zoumpourlis, V.C.; et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Vallerga, M.B.; Mansilla, S.F.; Federico, M.B.; Bertolin, A.P.; Gottifredi, V. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. USA 2015, 112, E6624–E6633. [Google Scholar] [CrossRef] [PubMed]
- Mazouzi, A.; Velimezi, G.; Loizou, J.I. DNA replication stress: Causes, resolution and disease. Exp. Cell Res. 2014, 329, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Jekimovs, C.; Bolderson, E.; Suraweera, A.; Adams, M.; O’Byrne, K.J.; Richard, D.J. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: The good, the bad, and the promising. Front. Oncol. 2014, 4, 86. [Google Scholar] [CrossRef] [PubMed]
- Beranek, D.T. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990, 231, 11–30. [Google Scholar] [CrossRef]
- Kondo, N.; Takahashi, A.; Mori, E.; Noda, T.; Su, X.; Ohnishi, K.; McKinnon, P.J.; Sakaki, T.; Nakase, H.; Ono, K.; et al. DNA ligase IV is a potential molecular target in ACNU sensitivity. Cancer Sci. 2010, 101, 1881–1885. [Google Scholar] [CrossRef] [PubMed]
- Brookes, P.; Lawley, P.D. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem. J. 1961, 80, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Lawley, P.D.; Brookes, P. The action of alkylating agents on deoxyribonucleic acid in relation to biological effects of the alkylating agents. Exp. Cell Res. 1963, 24 (Suppl. S9), 512–520. [Google Scholar] [CrossRef]
- Noll, D.M.; Mason, T.M.; Miller, P.S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006, 106, 277–301. [Google Scholar] [CrossRef] [PubMed]
- Schärer, O.D. DNA Interstrand Crosslinks: Natural and Drug-Induced DNA Adducts that Induce Unique Cellular Responses. ChemBioChem 2005, 6, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Lawley, P.D.; Phillips, D.H. DNA adducts from chemotherapeutic agents. Mutat. Res. 1996, 355, 13–40. [Google Scholar] [CrossRef]
- Bhuyan, B.K.; Scheidt, L.G.; Fraser, T.J. Cell cycle phase specificity of antitumor agents. Cancer Res. 1972, 32, 398–407. [Google Scholar] [PubMed]
- Glover, T.W.; Arlt, M.F.; Casper, A.M.; Durkin, S.G. Mechanisms of common fragile site instability. Hum. Mol. Genet. 2005, 14, R197–R205. [Google Scholar] [CrossRef] [PubMed]
- Koç, A.; Wheeler, L.J.; Mathews, C.K.; Merrill, G.F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 2004, 279, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Hsiang, Y.H.; Lihou, M.G.; Liu, L.F. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989, 49, 5077–5082. [Google Scholar] [PubMed]
- Deweese, J.E.; Osheroff, N. The DNA cleavage reaction of topoisomerase II: Wolf in sheep’s clothing. Nucleic Acids Res. 2009, 37, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjørås, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.J.; Schärer, O.D. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006, 106, 253–276. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair 2014, 19, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.-D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [PubMed]
- Bi, X. Mechanism of DNA damage tolerance. World J. Biol. Chem. 2015, 6, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Gómez-González, B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008, 9, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.J.; Cimprich, K.A. DNA damage tolerance: When it’s OK to make mistakes. Nat. Chem. Biol. 2009, 5, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, G.; Chen, J. DNA damage tolerance: A double-edged sword guarding the genome. Transl. Cancer Res. 2013, 2, 107–129. [Google Scholar] [PubMed]
- Saugar, I.; Ortiz-Bazán, M.Á.; Tercero, J.A. Tolerating DNA damage during eukaryotic chromosome replication. Exp. Cell Res. 2014, 329, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Deans, A.J.; West, S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Longerich, S.; Li, J.; Xiong, Y.; Sung, P.; Kupfer, G.M. Stress and DNA repair biology of the Fanconi anemia pathway. Blood 2014, 124, 2812–2819. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Mamrak, N.E.; Shimamura, A.; Howlett, N.G. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.D.; D’Andrea, A.D. The Fanconi Anemia/BRCA pathway: New faces in the crowd. Genes Dev. 2005, 19, 2925–2940. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H.; Hinz, J.M. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutat. Res. 2009, 668, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D. Ubiquitin family modifications and template switching. FEBS Lett. 2011, 585, 2810–2817. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Blackwell, S.; Lin, A.; Li, F.; Qin, Z.; Xiao, W. Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutat. Res. Rev. Mutat. Res. 2015, 764, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Q.; Jackson, D.A.; Blow, J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 2007, 21, 3331–3341. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Foiani, M.; Sogo, J.M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 2006, 21, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.M.; Göhler, T.; Luciani, M.G.; Oehlmann, M.; Ge, X.; Gartner, A.; Jackson, D.A.; Blow, J.J. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 2006, 173, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Elvers, I.; Johansson, F.; Groth, P.; Erixon, K.; Helleday, T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 2011, 39, 7049–7057. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, D.; Blow, J.J. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb. Perspect. Biol. 2012, 4, a012955. [Google Scholar] [CrossRef] [PubMed]
- De Piccoli, G.; Katou, Y.; Itoh, T.; Nakato, R.; Shirahige, K.; Labib, K. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol. Cell 2012, 45, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Tercero, J.A.; Diffley, J.F.X. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 2001, 412, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.; Cotta-Ramusino, C.; Pellicioli, A.; Liberi, G.; Plevani, P.; Muzi-Falconi, M.; Newlon, C.S.; Foiani, M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 2001, 412, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.A.; Bjergbaek, L.; Shimada, K.; Frei, C.; Gasser, S.M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003, 22, 4325–4336. [Google Scholar] [CrossRef] [PubMed]
- Ragland, R.L.; Patel, S.; Rivard, R.S.; Smith, K.; Peters, A.A.; Bielinsky, A.-K.; Brown, E.J. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013, 27, 2259–2273. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Budzowska, M.; Davies, S.L.; van Drunen, E.; Onizawa, H.; Beverloo, H.B.; Maas, A.; Essers, J.; Hickson, I.D.; Kanaar, R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007, 14, 1096–1104. [Google Scholar] [CrossRef] [PubMed]
- Forment, J.V.; Blasius, M.; Guerini, I.; Jackson, S.P. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS ONE 2011, 6, e23517. [Google Scholar] [CrossRef] [PubMed]
- Zellweger, R.; Dalcher, D.; Mutreja, K.; Berti, M.; Schmid, J.A.; Herrador, R.; Vindigni, A.; Lopes, M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015, 208, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Pacek, M.; Walter, J.C. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004, 23, 3667–3676. [Google Scholar] [CrossRef] [PubMed]
- Byun, T.S.; Pacek, M.; Yee, M.; Walter, J.C.; Cimprich, K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005, 19, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- MacDougall, C.A.; Byun, T.S.; Van, C.; Yee, M.; Cimprich, K.A. The structural determinants of checkpoint activation. Genes Dev. 2007, 21, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.A.; Cortez, D. ATR signalling: More than meeting at the fork. Biochem. J. 2011, 436, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Lucca, C.; Vanoli, F.; Cotta-Ramusino, C.; Pellicioli, A.; Liberi, G.; Haber, J.; Foiani, M. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 2004, 23, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Petermann, E.; Orta, M.L.; Issaeva, N.; Schultz, N.; Helleday, T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell 2010, 37, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Labib, K.; De Piccoli, G. Surviving chromosome replication: The many roles of the S-phase checkpoint pathway. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3554–3561. [Google Scholar] [CrossRef] [PubMed]
- Ozeri-Galai, E.; Schwartz, M.; Rahat, A.; Kerem, B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene 2008, 27, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Ammazzalorso, F.; Pirzio, L.M.; Bignami, M.; Franchitto, A.; Pichierri, P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010, 29, 3156–3169. [Google Scholar] [CrossRef] [PubMed]
- Bachrati, C.Z.; Hickson, I.D. RecQ helicases: Suppressors of tumorigenesis and premature aging. Biochem. J. 2003, 374, 577–606. [Google Scholar] [CrossRef] [PubMed]
- Hills, S.A.; Diffley, J.F.X. DNA replication and oncogene-induced replicative stress. Curr. Biol. 2014, 24, R435–R444. [Google Scholar] [CrossRef] [PubMed]
- Macheret, M.; Halazonetis, T.D. DNA replication stress as a hallmark of cancer. Annu. Rev. Pathol. 2015, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Murga, M.; Campaner, S.; Lopez-Contreras, A.J.; Toledo, L.I.; Soria, R.; Montaña, M.F.; D’Artista, L.; Schleker, T.; Guerra, C.; Garcia, E.; et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011, 18, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Marusyk, A.; Wheeler, L.J.; Mathews, C.K.; DeGregori, J. p53 mediates senescence-like arrest induced by chronic replicational stress. Mol. Cell. Biol. 2007, 27, 5336–5351. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Smolka, M.B.; Schimenti, J.C. Chronic DNA Replication Stress Reduces Replicative Lifespan of Cells by TRP53-Dependent, microRNA-Assisted MCM2–7 Downregulation. PLoS Genet. 2016, 12, e1005787. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Horejsí, Z.; Koed, K.; Krämer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehested, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. [Google Scholar] [CrossRef] [PubMed]
- O’Driscoll, M.; Ruiz-Perez, V.L.; Woods, C.G.; Jeggo, P.A.; Goodship, J.A. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003, 33, 497–501. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, P.J. ATM and ataxia telangiectasia. EMBO Rep. 2004, 5, 772–776. [Google Scholar] [CrossRef] [PubMed]
- DiGiovanna, J.J.; Kraemer, K.H. Shining a light on xeroderma pigmentosum. J. Investig. Dermatol. 2012, 132, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Callén, E.; Surrallés, J. Telomere dysfunction in genome instability syndromes. Mutat. Res. 2004, 567, 85–104. [Google Scholar] [CrossRef] [PubMed]
- Lauper, J.M.; Krause, A.; Vaughan, T.L.; Monnat, R.J. Spectrum and risk of neoplasia in Werner syndrome: A systematic review. PLoS ONE 2013, 8, e59709. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, K.A.; Gangloff, S.; Rothstein, R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010, 44, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; D’Andrea, A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012, 26, 1393–1408. [Google Scholar] [CrossRef] [PubMed]
- Joenje, H.; Patel, K.J. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2001, 2, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Larizza, L.; Roversi, G.; Volpi, L. Rothmund-Thomson syndrome. Orphanet J. Rare Dis. 2010, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Shamanna, R.A.; Keijzers, G.; Anand, R.; Rasmussen, L.J.; Cejka, P.; Croteau, D.L.; Bohr, V.A. RECQL4 Promotes DNA End Resection in Repair of DNA Double-Strand Breaks. Cell Rep. 2016, 16, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.; Vancamp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef] [PubMed]
- Todd, R.C.; Lippard, S.J. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009, 1, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Zamble, D.B.; Lippard, S.J. Cisplatin and DNA repair in cancer chemotherapy. Trends Biochem. Sci. 1995, 20, 435–439. [Google Scholar] [CrossRef]
- Available online: http://www.rcsb.org/pdb/explore/explore.do?structureId=3CO3 (accessed on 23 January 2017).
- Fichtinger-Schepman, A.M.; van der Veer, J.L.; den Hartog, J.H.; Lohman, P.H.; Reedijk, J. Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: Formation, identification, and quantitation. Biochemistry 1985, 24, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Harder, H.C.; Rosenberg, B. Inhibitory effects of anti-tumor platinum compounds on DNA, RNA and protein syntheses in mammalian cells in virtro. Int. J. Cancer 1970, 6, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A. Reevaluation of interaction of cis-dichloro(ethylenediamine)platinum(II) with DNA. Biochemistry 1986, 25, 3912–3915. [Google Scholar] [CrossRef] [PubMed]
- Sherman, S.E.; Gibson, D.; Wang, A.H.; Lippard, S.J. X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2(d(pGpG))]. Science 1985, 230, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Eastman, A. Separation and characterization of products resulting from the reaction of cis-diamminedichloroplatinum (II) with deoxyribonucleosides. Biochemistry 1982, 21, 6732–6736. [Google Scholar] [CrossRef] [PubMed]
- Desoize, B. Cancer and metals and metal compounds: Part I—Carcinogenesis. Crit. Rev. Oncol. Hematol. 2002, 42, 1–3. [Google Scholar] [CrossRef]
- Köberle, B.; Masters, J.R.; Hartley, J.A.; Wood, R.D. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr. Biol. 1999, 9, 273–276. [Google Scholar] [CrossRef]
- Borst, P.; Rottenberg, S.; Jonkers, J. How do real tumors become resistant to cisplatin? Cell Cycle 2008, 7, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Sedletska, Y.; Fourrier, L.; Malinge, J.-M. Modulation of MutS ATP-dependent functional activities by DNA containing a cisplatin compound lesion (base damage and mismatch). J. Mol. Biol. 2007, 369, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Clugston, C.; Burns, P.; Edlin, A.; Vasey, P.; Vojtĕsek, B.; Kaye, S.B. Increased accumulation of p53 protein in cisplatin-resistant ovarian cell lines. Int. J. Cancer 1993, 55, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Damsma, G.E.; Alt, A.; Brueckner, F.; Carell, T.; Cramer, P. Mechanism of transcriptional stalling at cisplatin-damaged DNA. Nat. Struct. Mol. Biol. 2007, 14, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Shimodaira, H.; Yoshioka-Yamashita, A.; Kolodner, R.D.; Wang, J.Y.J. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc. Natl. Acad. Sci. USA 2003, 100, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Aebi, S.; Kurdi-Haidar, B.; Gordon, R.; Cenni, B.; Zheng, H.; Fink, D.; Christen, R.D.; Boland, C.R.; Koi, M.; Fishel, R.; et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996, 56, 3087–3090. [Google Scholar] [PubMed]
- Alt, A.; Lammens, K.; Chiocchini, C.; Lammens, A.; Pieck, J.C.; Kuch, D.; Hopfner, K.-P.; Carell, T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 2007, 318, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Brozovic, A.; Ambriović-Ristov, A.; Osmak, M. The relationship between cisplatin-induced reactive oxygen species, glutathione, and BCL-2 and resistance to cisplatin. Crit. Rev. Toxicol. 2010, 40, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Splettstoesser, F.; Florea, A.-M.; Büsselberg, D. IP3 receptor antagonist, 2-APB, attenuates cisplatin induced Ca2+-influx in HeLa-S3 cells and prevents activation of calpain and induction of apoptosis. Br. J. Pharmacol. 2007, 151, 1176–1186. [Google Scholar] [CrossRef] [PubMed]
- Shamimi-Noori, S.; Yeow, W.-S.; Ziauddin, M.F.; Xin, H.; Tran, T.L.N.; Xie, J.; Loehfelm, A.; Patel, P.; Yang, J.; Schrump, D.S.; et al. Cisplatin enhances the antitumor effect of tumor necrosis factor-related apoptosis-inducing ligand gene therapy via recruitment of the mitochondria-dependent death signaling pathway. Cancer Gene Ther. 2008, 15, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Nishikawa, M.; Haque, A.M.; Hirose, M.; Mashimo, M.; Sato, E.; Inoue, M. Mitochondrial density determines the cellular sensitivity to cisplatin-induced cell death. Am. J. Physiol. Cell Physiol. 2005, 289, C1466–C1475. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, C.C.; Berberich, S.J. p53 binds to cisplatin-damaged DNA. Biochim. Biophys. Acta 2001, 1517, 392–397. [Google Scholar] [CrossRef]
- Kutuk, O.; Arisan, E.D.; Tezil, T.; Shoshan, M.C.; Basaga, H. Cisplatin overcomes Bcl-2-mediated resistance to apoptosis via preferential engagement of Bak: Critical role of Noxa-mediated lipid peroxidation. Carcinogenesis 2009, 30, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Hwang, J.-T.; Yun, H.; Chi, S.-G.; Lee, S.-J.; Kang, I.; Yoon, K.-S.; Choe, W.-J.; Kim, S.-S.; Ha, J. Inhibition of AMP-activated protein kinase sensitizes cancer cells to cisplatin-induced apoptosis via hyper-induction of p53. J. Biol. Chem. 2008, 283, 3731–3742. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Kaushal, V.; Haun, R.S.; Seth, R.; Shah, S.V.; Kaushal, G.P. Transcriptional activation of caspase-6 and -7 genes by cisplatin-induced p53 and its functional significance in cisplatin nephrotoxicity. Cell Death Differ. 2008, 15, 530–544. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Wei, Q.; Wang, J.; Du, Q.; Yu, J.; Zhang, L.; Dong, Z. Regulation of PUMA-alpha by p53 in cisplatin-induced renal cell apoptosis. Oncogene 2006, 25, 4056–4066. [Google Scholar] [CrossRef] [PubMed]
- Righetti, S.C.; Della Torre, G.; Pilotti, S.; Ménard, S.; Ottone, F.; Colnaghi, M.I.; Pierotti, M.A.; Lavarino, C.; Cornarotti, M.; Oriana, S.; et al. A comparative study of p53 gene mutations, protein accumulation, and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Cancer Res. 1996, 56, 689–693. [Google Scholar] [PubMed]
- Johnson, C.L.; Lu, D.; Huang, J.; Basu, A. Regulation of p53 stabilization by DNA damage and protein kinase C. Mol. Cancer Ther. 2002, 1, 861–867. [Google Scholar] [PubMed]
- Gong, J.G.; Costanzo, A.; Yang, H.Q.; Melino, G.; Kaelin, W.G.; Levrero, M.; Wang, J.Y. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature 1999, 399, 806–809. [Google Scholar] [PubMed]
- Preyer, M.; Shu, C.-W.; Wang, J.Y.J. Delayed activation of Bax by DNA damage in embryonic stem cells with knock-in mutations of the Abl nuclear localization signals. Cell Death Differ. 2007, 14, 1139–1148. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.K.C.; Yuan, Z.-M. c-Abl stabilizes p73 by a phosphorylation-augmented interaction. Cancer Res. 2003, 63, 3418–3424. [Google Scholar] [PubMed]
- Levy, D.; Adamovich, Y.; Reuven, N.; Shaul, Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol. Cell 2008, 29, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.V.; Dickman, M.J.; Whitmarsh, A.J. Regulation of p73-mediated apoptosis by c-Jun N-terminal kinase. Biochem. J. 2007, 405, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, J.; Ohmichi, M.; Kurachi, H.; Kanda, Y.; Hisamoto, K.; Nishio, Y.; Adachi, K.; Tasaka, K.; Kanzaki, T.; Murata, Y. Inhibition of BAD phosphorylation either at serine 112 via extracellular signal-regulated protein kinase cascade or at serine 136 via Akt cascade sensitizes human ovarian cancer cells to cisplatin. Cancer Res. 2000, 60, 5988–5994. [Google Scholar] [PubMed]
- Isonishi, S.; Andrews, P.A.; Howell, S.B. Increased sensitivity to cis-diamminedichloroplatinum(II) in human ovarian carcinoma cells in response to treatment with 12-O-tetradecanoylphorbol 13-acetate. J. Biol. Chem. 1990, 265, 3623–3627. [Google Scholar] [PubMed]
- Basu, A.; Teicher, B.A.; Lazo, J.S. Involvement of protein kinase C in phorbol ester-induced sensitization of HeLa cells to cis-diamminedichloroplatinum(II). J. Biol. Chem. 1990, 265, 8451–8457. [Google Scholar] [PubMed]
- Wang, X.; Dhalla, N.S. Modification of beta-adrenoceptor signal transduction pathway by genetic manipulation and heart failure. Mol. Cell. Biochem. 2000, 214, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Tu, H. Activation of ERK during DNA damage-induced apoptosis involves protein kinase Cdelta. Biochem. Biophys. Res. Commun. 2005, 334, 1068–1073. [Google Scholar] [CrossRef] [PubMed]
- Nowak, G. Protein kinase C-alpha and ERK1/2 mediate mitochondrial dysfunction, decreases in active Na+ transport, and cisplatin-induced apoptosis in renal cells. J. Biol. Chem. 2002, 277, 43377–43388. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pérez, I.; Benitah, S.A.; Martínez-Gomariz, M.; Lacal, J.C.; Perona, R. Cell stress and MEKK1-mediated c-Jun activation modulate NFκB activity and cell viability. Mol. Biol. Cell 2002, 13, 2933–2945. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Stroud, R.E.; Kaplan, B.S.; Leone, A.M.; Bavaria, J.E.; Gorman, J.H.; Gorman, R.C.; Ikonomidis, J.S. Differential protein kinase C isoform abundance in ascending aortic aneurysms from patients with bicuspid versus tricuspid aortic valves. Circulation 2007, 116, I144–I149. [Google Scholar] [CrossRef] [PubMed]
- Zanke, B.W.; Boudreau, K.; Rubie, E.; Winnett, E.; Tibbles, L.A.; Zon, L.; Kyriakis, J.; Liu, F.F.; Woodgett, J.R. The stress-activated protein kinase pathway mediates cell death following injury induced by cis-platinum, UV irradiation or heat. Curr. Biol. 1996, 6, 606–613. [Google Scholar] [CrossRef]
- Hernández Losa, J.; Parada Cobo, C.; Guinea Viniegra, J.; Sánchez-Arevalo Lobo, V.J.; Ramón y Cajal, S.; Sánchez-Prieto, R. Role of the p38 MAPK pathway in cisplatin-based therapy. Oncogene 2003, 22, 3998–4006. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Bourguignon, L.Y.W. Hyaluronan-CD44 promotes phospholipase C-mediated Ca2+ signaling and cisplatin resistance in head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 2006, 132, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Speelmans, G.; Staffhorst, R.W.; Versluis, K.; Reedijk, J.; de Kruijff, B. Cisplatin complexes with phosphatidylserine in membranes. Biochemistry 1997, 36, 10545–10550. [Google Scholar] [CrossRef] [PubMed]
- Huihui, Z.; Baohuai, W.; Youming, Z.; Kui, W. Calorimetric studies on actin polymerization and a comparison of the effects of cisplatin and transplatin. Thermochim. Acta 1995, 265, 31–38. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, Y.; Huang, Z.; Li, D.; Chen, X.; Cao, M.; Meng, Q.; Pang, H.; Sun, L.; Zhao, Y.; et al. miRNA-378 reverses chemoresistance to cisplatin in lung adenocarcinoma cells by targeting secreted clusterin. Sci. Rep. 2016, 6, 19455. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, H.; Liu, X.; Evans, B.R.; Medina, D.J.; Liu, C.-G.; Yang, J.-M. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem. Pharmacol. 2008, 76, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Vanas, V.; Haigl, B.; Stockhammer, V.; Sutterlüty-Fall, H. MicroRNA-21 Increases Proliferation and Cisplatin Sensitivity of Osteosarcoma-Derived Cells. PLoS ONE 2016, 11, e0161023. [Google Scholar] [CrossRef] [PubMed]
- Douple, E.B.; Richmond, R.C. Platinum complexes as radiosensitizers of hypoxic mammalian cells. Br. J. Cancer Suppl. 1978, 3, 98–102. [Google Scholar] [PubMed]
- Boeckman, H.J.; Trego, K.S.; Turchi, J.J. Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Mol. Cancer Res. 2005, 3, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, T.; Lippard, S.J. Telomere loss in cells treated with cisplatin. Proc. Natl. Acad. Sci. USA 1998, 95, 4219–4223. [Google Scholar] [CrossRef] [PubMed]
- Jordan, P.; Carmo-Fonseca, M. Cisplatin inhibits synthesis of ribosomal RNA in vivo. Nucleic Acids Res. 1998, 26, 2831–2836. [Google Scholar] [CrossRef] [PubMed]
- Tofilon, P.J.; Vines, C.M.; Baker, F.L.; Deen, D.F.; Brock, W.A. cis-Diamminedichloroplatinum(II)-induced sister chromatid exchange: An indicator of sensitivity and heterogeneity in primary human tumor cell cultures. Cancer Res. 1986, 46, 6156–6159. [Google Scholar] [PubMed]
- Berndtsson, M.; Hägg, M.; Panaretakis, T.; Havelka, A.M.; Shoshan, M.C.; Linder, S. Acute apoptosis by cisplatin requires induction of reactive oxygen species but is not associated with damage to nuclear DNA. Int. J. Cancer 2007, 120, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Rouette, A.; Parent, S.; Girouard, J.; Leblanc, V.; Asselin, E. Cisplatin increases B-cell-lymphoma-2 expression via activation of protein kinase C and Akt2 in endometrial cancer cells. Int. J. Cancer 2012, 130, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Damia, G.; Filiberti, L.; Vikhanskaya, F.; Carrassa, L.; Taya, Y.; D’incalci, M.; Broggini, M. Cisplatinum and taxol induce different patterns of p53 phosphorylation. Neoplasia 2001, 3, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Lützkendorf, J.; Wieduwild, E.; Nerger, K.; Lambrecht, N.; Schmoll, H.-J.; Müller-Tidow, C.; Müller, L.P. Resistance for Genotoxic Damage in Mesenchymal Stromal Cells Is Increased by Hypoxia but Not Generally Dependent on p53-Regulated Cell Cycle Arrest. PLoS ONE 2017, 12, e0169921. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, C.M.; Barry, M.A.; Eastman, A. Analysis of events associated with cell cycle arrest at G2 phase and cell death induced by cisplatin. J. Natl. Cancer Inst. 1990, 82, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Podratz, J.L.; Knight, A.M.; Ta, L.E.; Staff, N.P.; Gass, J.M.; Genelin, K.; Schlattau, A.; Lathroum, L.; Windebank, A.J. Cisplatin induced mitochondrial DNA damage in dorsal root ganglion neurons. Neurobiol. Dis. 2011, 41, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Karnitz, L.M. Cisplatin-induced DNA damage activates replication checkpoint signaling components that differentially affect tumor cell survival. Mol. Pharmacol. 2009, 76, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Bürkle, A.; Chen, G.; Küpper, J.H.; Grube, K.; Zeller, W.J. Increased poly(ADP-ribosyl)ation in intact cells by cisplatin treatment. Carcinogenesis 1993, 14, 559–561. [Google Scholar] [CrossRef] [PubMed]
- Jennerwein, M.; Andrews, P.A. Drug accumulation and DNA platination in cells exposed to aquated cisplatin species. Cancer Lett. 1994, 81, 215–220. [Google Scholar] [CrossRef]
- Shirazi, F.H.; Molepo, J.M.; Stewart, D.J.; Ng, C.E.; Raaphorst, G.P.; Goel, R. Cytotoxicity, accumulation, and efflux of cisplatin and its metabolites in human ovarian carcinoma cells. Toxicol. Appl. Pharmacol. 1996, 140, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Telma, K.A.; Chang, K.-E.; Lee, T.D.; Madigan, J.P.; Lloyd, J.R.; Goldlust, I.S.; Hoeschele, J.D.; Gottesman, M.M. Say no to DMSO: Dimethylsulfoxide inactivates cisplatin, carboplatin, and other platinum complexes. Cancer Res. 2014, 74, 3913–3922. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.webcitation.org/6mEemW8DM (accessed on 23 November 2016).
- Massart, C.; Le Tellier, C.; Gibassier, J.; Leclech, G.; Nicol, M. Modulation by dimethyl sulphoxide of the toxicity induced by cis-diamminedichloroplatinum in cultured thyrocytes. Toxicol. Vitro 1993, 7, 87–94. [Google Scholar] [CrossRef]
- Wiltshaw, E.; Subramarian, S.; Alexopoulos, C.; Barker, G.H. Cancer of the ovary: A summary of experience with cis-dichlorodiammineplatinum(II) at the Royal Marsden Hospital. Cancer Treat. Rep. 1979, 63, 1545–1548. [Google Scholar] [PubMed]
- Galanski, M. Recent developments in the field of anticancer platinum complexes. Recent Pat. Anticancer Drug Discov. 2006, 1, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Lebwohl, D.; Canetta, R. Clinical development of platinum complexes in cancer therapy: An historical perspective and an update. Eur. J. Cancer 1998, 34, 1522–1534. [Google Scholar] [CrossRef]
- Baetz, T.; Belch, A.; Couban, S.; Imrie, K.; Yau, J.; Myers, R.; Ding, K.; Paul, N.; Shepherd, L.; Iglesias, J.; et al. Gemcitabine, dexamethasone and cisplatin is an active and non-toxic chemotherapy regimen in relapsed or refractory Hodgkin’s disease: A phase II study by the National Cancer Institute of Canada Clinical Trials Group. Ann. Oncol. 2003, 14, 1762–1767. [Google Scholar] [CrossRef] [PubMed]
- Crump, M.; Baetz, T.; Couban, S.; Belch, A.; Marcellus, D.; Howson-Jan, K.; Imrie, K.; Myers, R.; Adams, G.; Ding, K.; et al. Gemcitabine, dexamethasone, and cisplatin in patients with recurrent or refractory aggressive histology B-cell non-Hodgkin lymphoma: A Phase II study by the National Cancer Institute of Canada Clinical Trials Group (NCIC-CTG). Cancer 2004, 101, 1835–1842. [Google Scholar] [CrossRef] [PubMed]
- Pearson, A.D.J.; Pinkerton, C.R.; Lewis, I.J.; Imeson, J.; Ellershaw, C.; Machin, D.; European Neuroblastoma Study Group; Children’s Cancer and Leukaemia Group (CCLG formerly United Kingdom Children’s Cancer Study Group). High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: A randomised trial. Lancet Oncol. 2008, 9, 247–256. [Google Scholar] [PubMed]
- Reichardt, P. The treatment of uterine sarcomas. Ann. Oncol. 2012, 23 (Suppl. S10), x151–x157. [Google Scholar] [CrossRef] [PubMed]
- Dadacaridou, M.; Papanicolaou, X.; Maltesas, D.; Megalakaki, C.; Patos, P.; Panteli, K.; Repousis, P.; Mitsouli-Mentzikof, C. Dexamethasone, cyclophosphamide, etoposide and cisplatin (DCEP) for relapsed or refractory multiple myeloma patients. J. BUON 2007, 12, 41–44. [Google Scholar] [PubMed]
- Glover, D.; Glick, J.H.; Weiler, C.; Fox, K.; Guerry, D. WR-2721 and high-dose cisplatin: An active combination in the treatment of metastatic melanoma. J. Clin. Oncol. 1987, 5, 574–578. [Google Scholar] [PubMed]
- Berghmans, T.; Paesmans, M.; Lalami, Y.; Louviaux, I.; Luce, S.; Mascaux, C.; Meert, A.P.; Sculier, J.P. Activity of chemotherapy and immunotherapy on malignant mesothelioma: A systematic review of the literature with meta-analysis. Lung Cancer 2002, 38, 111–121. [Google Scholar] [CrossRef]
- Hanada, K.; Nishijima, K.; Ogata, H.; Atagi, S.; Kawahara, M. Population pharmacokinetic analysis of cisplatin and its metabolites in cancer patients: Possible misinterpretation of covariates for pharmacokinetic parameters calculated from the concentrations of unchanged cisplatin, ultrafiltered platinum and total platinum. Jpn. J. Clin. Oncol. 2001, 31, 179–184. [Google Scholar] [PubMed]
- Daugaard, G.; Abildgaard, U. Cisplatin nephrotoxicity. A review. Cancer Chemother. Pharmacol. 1989, 25, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nagai, N.; Kinoshita, M.; Ogata, H.; Tsujino, D.; Wada, Y.; Someya, K.; Ohno, T.; Masuhara, K.; Tanaka, Y.; Kato, K.; et al. Relationship between pharmacokinetics of unchanged cisplatin and nephrotoxicity after intravenous infusions of cisplatin to cancer patients. Cancer Chemother. Pharmacol. 1996, 39, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Kartalou, M.; Essigmann, J.M. Mechanisms of resistance to cisplatin. Mutat. Res. 2001, 478, 23–43. [Google Scholar] [CrossRef]
- Olszewski, U.; Hamilton, G. A better platinum-based anticancer drug yet to come? Anticancer Agents Med. Chem. 2010, 10, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Dieras, V.; Girre, V.; Guilhaume, M.-N.; Laurence, V.; Mignot, L. Oxaliplatin and ovarian cancer. Bull. Cancer 2006, 93 (Suppl. S1), S35–S39. [Google Scholar] [PubMed]
- Ganjavi, H.; Gee, M.; Narendran, A.; Parkinson, N.; Krishnamoorthy, M.; Freedman, M.H.; Malkin, D. Adenovirus-mediated p53 gene therapy in osteosarcoma cell lines: Sensitization to cisplatin and doxorubicin. Cancer Gene Ther. 2006, 13, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Michels, J.; Vitale, I.; Senovilla, L.; Enot, D.P.; Garcia, P.; Lissa, D.; Olaussen, K.A.; Brenner, C.; Soria, J.-C.; Castedo, M.; et al. Synergistic interaction between cisplatin and PARP inhibitors in non-small cell lung cancer. Cell Cycle 2013, 12, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Balmaña, J.; Tung, N.M.; Isakoff, S.J.; Graña, B.; Ryan, P.D.; Saura, C.; Lowe, E.S.; Frewer, P.; Winer, E.; Baselga, J.; et al. Phase I trial of olaparib in combination with cisplatin for the treatment of patients with advanced breast, ovarian and other solid tumors. Ann. Oncol. 2014, 25, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, C.M.; Eastman, A. Influence of cis-diamminedichloroplatinum(II) on DNA synthesis and cell cycle progression in excision repair proficient and deficient Chinese hamster ovary cells. Cancer Res. 1988, 48, 6703–6707. [Google Scholar] [PubMed]
- Vichi, P.; Coin, F.; Renaud, J.P.; Vermeulen, W.; Hoeijmakers, J.H.; Moras, D.; Egly, J.M. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997, 16, 7444–7456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullinane, C.; Mazur, S.J.; Essigmann, J.M.; Phillips, D.R.; Bohr, V.A. Inhibition of RNA polymerase II transcription in human cell extracts by cisplatin DNA damage. Biochemistry 1999, 38, 6204–6212. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, A.; Shah, P.P.; Rai, S.N.; Panguluri, S.K.; Kakar, S.S. MicroRNA signature of cis-platin resistant vs. cis-platin sensitive ovarian cancer cell lines. J. Ovarian Res. 2011, 4, 17. [Google Scholar] [PubMed]
- Ciarimboli, G.; Ludwig, T.; Lang, D.; Pavenstädt, H.; Koepsell, H.; Piechota, H.-J.; Haier, J.; Jaehde, U.; Zisowsky, J.; Schlatter, E. Cisplatin nephrotoxicity is critically mediated via the human organic cation transporter 2. Am. J. Pathol. 2005, 167, 1477–1484. [Google Scholar] [CrossRef]
- Gressette, M.; Vérillaud, B.; Jimenez-Pailhès, A.-S.; Lelièvre, H.; Lo, K.-W.; Ferrand, F.-R.; Gattolliat, C.-H.; Jacquet-Bescond, A.; Kraus-Berthier, L.; Depil, S.; et al. Treatment of Nasopharyngeal Carcinoma Cells with the Histone-Deacetylase Inhibitor Abexinostat: Cooperative Effects with Cis-platin and Radiotherapy on Patient-Derived Xenografts. PLoS ONE 2014, 9, e91325. [Google Scholar] [CrossRef] [PubMed]
- Rout, S.R.; Behera, B.; Maiti, T.K.; Mohapatra, S. Multifunctional magnetic calcium phosphate nanoparticles for targeted platin delivery. Dalton Trans. 2012, 41, 10777–10783. [Google Scholar] [CrossRef] [PubMed]
- Kitao, H.; Takata, M. Fanconi anemia: A disorder defective in the DNA damage response. Int. J. Hematol. 2011, 93, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.; Kothandapani, A.; Zhitkovich, A.; Sobol, R.W.; Patrick, S.M. Role of mismatch repair proteins in the processing of cisplatin interstrand cross-links. DNA Repair 2015, 35, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.H.; Kuchta, R.D. DNA polymerase epsilon: Aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry 1993, 32, 8568–8574. [Google Scholar] [CrossRef] [PubMed]
- Pedrali-Noy, G.; Spadari, S.; Miller-Faurès, A.; Miller, A.O.; Kruppa, J.; Koch, G. Synchronization of HeLa cell cultures by inhibition of DNA polymerase alpha with aphidicolin. Nucleic Acids Res. 1980, 8, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Baranovskiy, A.G.; Babayeva, N.D.; Suwa, Y.; Gu, J.; Pavlov, Y.I.; Tahirov, T.H. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 2014, 42, 14013–14021. [Google Scholar] [CrossRef] [PubMed]
- Spadari, S.; Pedrali-Noy, G.; Falaschi, M.C.; Ciarrocchi, G. Control of DNA replication and cell proliferation in eukaryotes by aphidicolin. Toxicol. Pathol. 1984, 12, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.rcsb.org/pdb/explore/explore.do?structureId=4Q5V) (accessed on 23 January 2017).
- Chang, D.J.; Lupardus, P.J.; Cimprich, K.A. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J. Biol. Chem. 2006, 281, 32081–32088. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, G.R. Chromosomal fragile sites. Genet. Anal. Tech. Appl. 1991, 8, 161–166. [Google Scholar] [CrossRef]
- Shiraishi, T.; Druck, T.; Mimori, K.; Flomenberg, J.; Berk, L.; Alder, H.; Miller, W.; Huebner, K.; Croce, C.M. Sequence conservation at human and mouse orthologous common fragile regions, FRA3B/FHIT and Fra14A2/Fhit. Proc. Natl. Acad. Sci. USA 2001, 98, 5722–5727. [Google Scholar] [CrossRef] [PubMed]
- Hellman, A.; Zlotorynski, E.; Scherer, S.W.; Cheung, J.; Vincent, J.B.; Smith, D.I.; Trakhtenbrot, L.; Kerem, B. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 2002, 1, 89–97. [Google Scholar] [CrossRef]
- Durkin, S.G.; Ragland, R.L.; Arlt, M.F.; Mulle, J.G.; Warren, S.T.; Glover, T.W. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl. Acad. Sci. USA 2008, 105, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.G.; Hill, R.P. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008, 8, 180–192. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, J.T.; Schlegel, R.; Wehr, C.M.; Alperin, P.; Ames, B.N. Cytogenetic damage induced by folate deficiency in mice is enhanced by caffeine. Proc. Natl. Acad. Sci. USA 1990, 87, 9962–9965. [Google Scholar] [CrossRef] [PubMed]
- Koundrioukoff, S.; Carignon, S.; Técher, H.; Letessier, A.; Brison, O.; Debatisse, M. Stepwise activation of the ATR signaling pathway upon increasing replication stress impacts fragile site integrity. PLoS Genet. 2013, 9, e1003643. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 2011, 44, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre’, M.; Nuciforo, P.G.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef]
- Cha, R.S.; Kleckner, N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 2002, 297, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Arlt, M.F.; Mulle, J.G.; Schaibley, V.M.; Ragland, R.L.; Durkin, S.G.; Warren, S.T.; Glover, T.W. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am. J. Hum. Genet. 2009, 84, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Hardt, N.; Pedrali-Noy, G.; Focher, F.; Spadari, S. Aphidicolin does not inhibit DNA repair synthesis in ultraviolet-irradiated HeLa cells. A radioautographic study. Biochem. J. 1981, 199, 453–455. [Google Scholar] [CrossRef] [PubMed]
- Pedrali-Noy, G.; Belvedere, M.; Crepaldi, T.; Focher, F.; Spadari, S. Inhibition of DNA replication and growth of several human and murine neoplastic cells by aphidicolin without detectable effect upon synthesis of immunoglobulins and HLA antigens. Cancer Res. 1982, 42, 3810–3813. [Google Scholar] [PubMed]
- Gera, J.F.; Fady, C.; Gardner, A.; Jacoby, F.J.; Briskin, K.B.; Lichtenstein, A. Inhibition of DNA repair with aphidicolin enhances sensitivity of targets to tumor necrosis factor. J. Immunol. 1993, 151, 3746–3757. [Google Scholar] [PubMed]
- Waters, R. Aphidicolin: An inhibitor of DNA repair in human fibroblasts. Carcinogenesis 1981, 2, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Stewart, J.; Price, C.M. Human CST abundance determines recovery from diverse forms of DNA damage and replication stress. Cell Cycle 2014, 13, 3488–3498. [Google Scholar] [CrossRef] [PubMed]
- Yeo, J.E.; Lee, E.H.; Hendrickson, E.A.; Sobeck, A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum. Mol. Genet. 2014, 23, 3695–3705. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, I.; Stroik, D.R.; Sobeck, A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol. Cell. Biol. 2014, 34, 3939–3954. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.M.; Green, S.L.; Giaccia, A.J. Comparison of hypoxia-induced replication arrest with hydroxyurea and aphidicolin-induced arrest. Mutat. Res. 2003, 532, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Lacroix, F.B.; Margolis, R.L. Prolonged arrest of mammalian cells at the G1/S boundary results in permanent S phase stasis. J. Cell Sci. 2002, 115, 2829–2838. [Google Scholar] [PubMed]
- Basile, G.; Leuzzi, G.; Pichierri, P.; Franchitto, A. Checkpoint-dependent and independent roles of the Werner syndrome protein in preserving genome integrity in response to mild replication stress. Nucleic Acids Res. 2014, 42, 12628–12639. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.H.; Dexheimer, T.S.; Rosenthal, A.S.; Chu, W.K.; Singh, D.K.; Mosedale, G.; Bachrati, C.Z.; Schultz, L.; Sakurai, M.; Savitsky, P.; et al. A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 2013, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.; Wiedner, M.; Velimezi, G.; Prochazkova, J.; Owusu, M.; Bauer, S.; Loizou, J.I. ATMIN is required for the ATM-mediated signaling and recruitment of 53BP1 to DNA damage sites upon replication stress. DNA Repair 2014, 24, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Sasanuma, H.; Yamamoto, K.N.; Harada, H.; Kurosawa, A.; Adachi, N.; Omura, M.; Hiraoka, M.; Takeda, S.; Hirota, K. Interference in DNA replication can cause mitotic chromosomal breakage unassociated with double-strand breaks. PLoS ONE 2013, 8, e60043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beresova, L.; Vesela, E.; Chamrad, I.; Voller, J.; Yamada, M.; Furst, T.; Lenobel, R.; Chroma, K.; Gursky, J.; Krizova, K.; et al. Role of DNA Repair Factor Xeroderma Pigmentosum Protein Group C in Response to Replication Stress As Revealed by DNA Fragile Site Affinity Chromatography and Quantitative Proteomics. J. Proteome Res. 2016, 15, 4505–4517. [Google Scholar] [CrossRef] [PubMed]
- Janson, C.; Nyhan, K.; Murnane, J.P. Replication Stress and Telomere Dysfunction Are Present in Cultured Human Embryonic Stem Cells. Cytogenet. Genome Res. 2015, 146, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Miron, K.; Golan-Lev, T.; Dvir, R.; Ben-David, E.; Kerem, B. Oncogenes create a unique landscape of fragile sites. Nat. Commun. 2015, 6, 7094. [Google Scholar] [CrossRef] [PubMed]
- Murfuni, I.; De Santis, A.; Federico, M.; Bignami, M.; Pichierri, P.; Franchitto, A. Perturbed replication induced genome wide or at common fragile sites is differently managed in the absence of WRN. Carcinogenesis 2012, 33, 1655–1663. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, T.; Magdalou, I.; Barascu, A.; Técher, H.; Debatisse, M.; Lopez, B.S. Spontaneous slow replication fork progression elicits mitosis alterations in homologous recombination-deficient mammalian cells. Proc. Natl. Acad. Sci. USA 2014, 111, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.google.cz/url?sa=t&rct=j&q=&esrc=s&source=web&cd=3&cad=rja&uact=8&ved=0ahUKEwibvoX5jL_QAhULVSwKHQfLCXwQFggsMAI&url=https%3A%2F%2Fwww.sigmaaldrich.com%2Fcontent%2Fdam%2Fsigma-aldrich%2Fdocs%2FSigma%2FDatasheet%2F6%2Fa0781dat.pdf&usg=AFQjCNEPSqAi (accessed on 23 November 2016).
- Sessa, C.; Zucchetti, M.; Davoli, E.; Califano, R.; Cavalli, F.; Frustaci, S.; Gumbrell, L.; Sulkes, A.; Winograd, B.; D’Incalci, M. Phase I and clinical pharmacological evaluation of aphidicolin glycinate. J. Natl. Cancer Inst. 1991, 83, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Edelson, R.E.; Gorycki, P.D.; MacDonald, T.L. The mechanism of aphidicolin bioinactivation by rat liver in vitro systems. Xenobiotica 1990, 20, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.B.; Krogh, R.; Magalhaes, L.G.; Andricopulo, A.D.; Pupo, M.T.; Emery, F.S. Semisynthesis of new aphidicolin derivatives with high activity against Trypanosoma cruzi. Bioorg. Med. Chem. Lett. 2016, 26, 1205–1208. [Google Scholar] [CrossRef] [PubMed]
- Glover, T.W.; Berger, C.; Coyle, J.; Echo, B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984, 67, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Kurose, A.; Tanaka, T.; Huang, X.; Traganos, F.; Darzynkiewicz, Z. Synchronization in the cell cycle by inhibitors of DNA replication induces histone H2AX phosphorylation: An indication of DNA damage. Cell Prolif. 2006, 39, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Trenz, K.; Smith, E.; Smith, S.; Costanzo, V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J. 2006, 25, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Krakoff, I.H.; Brown, N.C.; Reichard, P. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res. 1968, 28, 1559–1565. [Google Scholar] [PubMed]
- Reichard, P. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 1988, 57, 349–374. [Google Scholar] [CrossRef] [PubMed]
- Håkansson, P.; Hofer, A.; Thelander, L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 2006, 281, 7834–7841. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Uhlin, U.; Ramaswamy, S.; Ekberg, M.; Regnström, K.; Sjöberg, B.M.; Eklund, H. Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure 1997, 5, 1077–1092. [Google Scholar] [CrossRef]
- Bianchi, V.; Pontis, E.; Reichard, P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 1986, 261, 16037–16042. [Google Scholar] [PubMed]
- Skog, S.; Tribukait, B.; Wallström, B.; Eriksson, S. Hydroxyurea-induced cell death as related to cell cycle in mouse and human T-lymphoma cells. Cancer Res. 1987, 47, 6490–6493. [Google Scholar] [PubMed]
- Akerblom, L. Azidocytidine is incorporated into RNA of 3T6 mouse fibroblasts. FEBS Lett. 1985, 193, 203–207. [Google Scholar] [CrossRef]
- Anglana, M.; Apiou, F.; Bensimon, A.; Debatisse, M. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 2003, 114, 385–394. [Google Scholar] [CrossRef]
- Barlow, J.H.; Faryabi, R.B.; Callén, E.; Wong, N.; Malhowski, A.; Chen, H.T.; Gutierrez-Cruz, G.; Sun, H.-W.; McKinnon, P.; Wright, G.; et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 2013, 152, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Lönn, U.; Lönn, S. Extensive regions of single-stranded DNA in aphidicolin-treated melanoma cells. Biochemistry 1988, 27, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Recolin, B.; Van der Laan, S.; Maiorano, D. Role of replication protein A as sensor in activation of the S-phase checkpoint in Xenopus egg extracts. Nucleic Acids Res. 2012, 40, 3431–3442. [Google Scholar] [CrossRef] [PubMed]
- Arlt, M.F.; Ozdemir, A.C.; Birkeland, S.R.; Wilson, T.E.; Glover, T.W. Hydroxyurea induces de novo copy number variants in human cells. Proc. Natl. Acad. Sci. USA 2011, 108, 17360–17365. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.-E.; Facca, C.; Fatmi, Z.; Baïlle, D.; Bénakli, S.; Vernis, L. DNA replication inhibitor hydroxyurea alters Fe–S centers by producing reactive oxygen species in vivo. Sci. Rep. 2016, 6, 29361. [Google Scholar] [CrossRef] [PubMed]
- Szikriszt, B.; Póti, Á.; Pipek, O.; Krzystanek, M.; Kanu, N.; Molnár, J.; Ribli, D.; Szeltner, Z.; Tusnády, G.E.; Csabai, I.; et al. A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol. 2016, 17, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistrik, M.; Oplustilova, L.; Lukas, J.; Bartek, J. Low-dose DNA damage and replication stress responses quantified by optimized automated single-cell image analysis. Cell Cycle 2009, 8, 2592–2599. [Google Scholar] [CrossRef] [PubMed]
- Ohouo, P.Y.; Bastos de Oliveira, F.M.; Liu, Y.; Ma, C.J.; Smolka, M.B. DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9. Nature 2013, 493, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Morafraile, E.C.; Diffley, J.F.X.; Tercero, J.A.; Segurado, M. Checkpoint-dependent RNR induction promotes fork restart after replicative stress. Sci. Rep. 2015, 5, 7886. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-S.; Kim, S.-K.; Hromas, R.; Lee, S.-H. The SET Domain Is Essential for Metnase Functions in Replication Restart and the 5’ End of SS-Overhang Cleavage. PLoS ONE 2015, 10, e0139418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Xu, X.; Dimitriadis, E.K.; Lahusen, T.; Deng, C.-X. “DNA Binding Region” of BRCA1 Affects Genetic Stability through modulating the Intra-S-Phase Checkpoint. Int. J. Biol. Sci. 2016, 12, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Yarden, R.I.; Metsuyanim, S.; Pickholtz, I.; Shabbeer, S.; Tellio, H.; Papa, M.Z. BRCA1-dependent Chk1 phosphorylation triggers partial chromatin disassociation of phosphorylated Chk1 and facilitates S-phase cell cycle arrest. Int. J. Biochem. Cell Biol. 2012, 44, 1761–1769. [Google Scholar] [CrossRef] [PubMed]
- Awate, S.; De Benedetti, A. TLK1B mediated phosphorylation of Rad9 regulates its nuclear/cytoplasmic localization and cell cycle checkpoint. BMC Mol. Biol. 2016, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Ahlskog, J.K.; Larsen, B.D.; Achanta, K.; Sørensen, C.S. ATM/ATR-mediated phosphorylation of PALB2 promotes RAD51 function. EMBO Rep. 2016, 17, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Molina, B.; Marchetti, F.; Gómez, L.; Ramos, S.; Torres, L.; Ortiz, R.; Altamirano-Lozano, M.; Carnevale, A.; Frias, S. Hydroxyurea induces chromosomal damage in G2 and enhances the clastogenic effect of mitomycin C in Fanconi anemia cells. Environ. Mol. Mutagen. 2015, 56, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Croke, M.; Neumann, M.A.; Grotsky, D.A.; Kreienkamp, R.; Yaddanapudi, S.C.; Gonzalo, S. Differences in 53BP1 and BRCA1 regulation between cycling and non-cycling cells. Cell Cycle 2013, 12, 3629–3639. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Watanabe, K.; Mistrik, M.; Vesela, E.; Protivankova, I.; Mailand, N.; Lee, M.; Masai, H.; Lukas, J.; Bartek, J. ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 2013, 27, 2459–2472. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Kim, T.M.; Son, M.Y.; Kim, S.-A.; Holland, C.L.; Tateishi, S.; Kim, D.H.; Yew, P.R.; Montagna, C.; Dumitrache, L.C.; et al. Two replication fork maintenance pathways fuse inverted repeats to rearrange chromosomes. Nature 2013, 501, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Lou, T.-F.; Singh, M.; Mackie, A.; Li, W.; Pace, B.S. Hydroxyurea generates nitric oxide in human erythroid cells: Mechanisms for gamma-globin gene activation. Exp. Biol. Med. 2009, 234, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, I.; Yanakieva, I.; Peycheva, M.; Gospodinov, A.; Anachkova, B. The mammalian INO80 chromatin remodeling complex is required for replication stress recovery. Nucleic Acids Res. 2014, 42, 9074–9086. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Choi, H.S.; Jeong, J.S.; Han, J.Y.; Kim, I.H. Involvement of p38 kinase in hydroxyurea-induced differentiation of K562 cells. Cell Growth Differ. 2001, 12, 481–486. [Google Scholar] [PubMed]
- Barthelemy, J.; Hanenberg, H.; Leffak, M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2016, 44, 6803–6816. [Google Scholar] [CrossRef] [PubMed]
- Kunnev, D.; Rusiniak, M.E.; Kudla, A.; Freeland, A.; Cady, G.K.; Pruitt, S.C. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene 2010, 29, 3630–3638. [Google Scholar] [CrossRef] [PubMed]
- Da Guarda, C.C.; Santiago, R.P.; Pitanga, T.N.; Santana, S.S.; Zanette, D.L.; Borges, V.M.; Goncalves, M.S. Heme changes HIF-α, eNOS and nitrite production in HUVECs after simvastatin, HU, and ascorbic acid therapies. Microvasc. Res. 2016, 106, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Leitch, C.; Osdal, T.; Andresen, V.; Molland, M.; Kristiansen, S.; Nguyen, X.N.; Bruserud, Ø.; Gjertsen, B.T.; McCormack, E. Hydroxyurea synergizes with valproic acid in wild-type p53 acute myeloid leukaemia. Oncotarget 2016, 7, 8105–8118. [Google Scholar] [PubMed]
- Liu, K.; Graves, J.D.; Scott, J.D.; Li, R.; Lin, W.-C. Akt switches TopBP1 function from checkpoint activation to transcriptional regulation through phosphoserine binding-mediated oligomerization. Mol. Cell. Biol. 2013, 33, 4685–4700. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.google.cz/url?sa=t&rct=j&q=&esrc=s&source=web&cd=3&cad=rja&uact=8&ved=0ahUKEwiVt4Lulb_QAhUBGSwKHbcOB_kQFggsMAI&url=https%3A%2F%2Fwww.sigmaaldrich.com%2Fcontent%2Fdam%2Fsigma-aldrich%2Fdocs%2FSigma%2FProduct_Information_Sheet%2F2%2Fh8627pis.pdf& (accessed on 23 January 2017).
- Segal, J.B.; Strouse, J.J.; Beach, M.C.; Haywood, C.; Witkop, C.; Park, H.; Wilson, R.F.; Bass, E.B.; Lanzkron, S. Hydroxyurea for the Treatment of Sickle Cell Disease; Evidence Reports/Technology Assessments; Agency for Healthcare Research and Quality (US): Rockville, MD, USA, 2008; pp. 1–95.
- Kühr, T.; Burgstaller, S.; Apfelbeck, U.; Linkesch, W.; Seewann, H.; Fridrik, M.; Michlmayr, G.; Krieger, O.; Lutz, D.; Lin, W.; et al. A randomized study comparing interferon (IFNα) plus low-dose cytarabine and interferon plus hydroxyurea (HU) in early chronic-phase chronic myeloid leukemia (CML). Leuk. Res. 2003, 27, 405–411. [Google Scholar] [CrossRef]
- Aruch, D.; Mascarenhas, J. Contemporary approach to essential thrombocythemia and polycythemia vera. Curr. Opin. Hematol. 2016, 23, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Barbui, T.; Finazzi, M.C.; Finazzi, G. Front-line therapy in polycythemia vera and essential thrombocythemia. Blood Rev. 2012, 26, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Benito, J.M.; López, M.; Lozano, S.; Ballesteros, C.; González-Lahoz, J.; Soriano, V. Hydroxyurea exerts an anti-proliferative effect on T cells but has no direct impact on cellular activation. Clin. Exp. Immunol. 2007, 149, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Gurberg, J.; Bouganim, N.; Shenouda, G.; Zeitouni, A. A case of recurrent anaplastic meningioma of the skull base with radiologic response to hydroxyurea. J. Neurol. Surg. Rep. 2014, 75, e52–e55. [Google Scholar] [CrossRef] [PubMed]
- Kiladjian, J.-J.; Chevret, S.; Dosquet, C.; Chomienne, C.; Rain, J.-D. Treatment of polycythemia vera with hydroxyurea and pipobroman: Final results of a randomized trial initiated in 1980. J. Clin. Oncol. 2011, 29, 3907–3913. [Google Scholar] [CrossRef] [PubMed]
- Charache, S.; Barton, F.B.; Moore, R.D.; Terrin, M.L.; Steinberg, M.H.; Dover, G.J.; Ballas, S.K.; McMahon, R.P.; Castro, O.; Orringer, E.P. Hydroxyurea and sickle cell anemia. Clinical utility of a myelosuppressive “switching” agent. The Multicenter Study of Hydroxyurea in Sickle Cell Anemia. Medicine 1996, 75, 300–326. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, M.H.; McCarthy, W.F.; Castro, O.; Ballas, S.K.; Armstrong, F.D.; Smith, W.; Ataga, K.; Swerdlow, P.; Kutlar, A.; DeCastro, L.; et al. The risks and benefits of long-term use of hydroxyurea in sickle cell anemia: A 17.5 year follow-up. Am. J. Hematol. 2010, 85, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Darzynkiewicz, Z.; Halicka, H.D.; Zhao, H.; Podhorecka, M. Cell synchronization by inhibitors of DNA replication induces replication stress and DNA damage response: Analysis by flow cytometry. Methods Mol. Biol. 2011, 761, 85–96. [Google Scholar] [PubMed]
- Fugger, K.; Mistrik, M.; Danielsen, J.R.; Dinant, C.; Falck, J.; Bartek, J.; Lukas, J.; Mailand, N. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 2009, 186, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Lim, C.-S. Differential roles of XRCC2 in homologous recombinational repair of stalled replication forks. J. Cell. Biochem. 2005, 95, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Brose, R.D.; Shin, G.; McGuinness, M.C.; Schneidereith, T.; Purvis, S.; Dong, G.X.; Keefer, J.; Spencer, F.; Smith, K.D. Activation of the stress proteome as a mechanism for small molecule therapeutics. Hum. Mol. Genet. 2012, 21, 4237–4252. [Google Scholar] [CrossRef] [PubMed]
- Adragna, N.C.; Fonseca, P.; Lauf, P.K. Hydroxyurea affects cell morphology, cation transport, and red blood cell adhesion in cultured vascular endothelial cells. Blood 1994, 83, 553–560. [Google Scholar] [PubMed]
- Wall, M.E.; Wani, M.C.; Cook, C.E.; Palmer, K.H.; McPhail, A.T.; Sim, G.A. Plant Antitumor Agents. I. The Isolation and Structure of Camptothecin, a Novel Alkaloidal Leukemia and Tumor Inhibitor from Camptotheca acuminata1,2. J. Am. Chem. Soc. 1966, 88, 3888–3890. [Google Scholar] [CrossRef]
- Gupta, M.; Fujimori, A.; Pommier, Y. Eukaryotic DNA topoisomerases I. Biochim. Biophys. Acta 1995, 1262, 1–14. [Google Scholar] [CrossRef]
- Champoux, J.J. Mechanism of the reaction catalyzed by the DNA untwisting enzyme: Attachment of the enzyme to 3′-terminus of the nicked DNA. J. Mol. Biol. 1978, 118, 441–446. [Google Scholar] [CrossRef]
- Available online: http://www.rcsb.org/pdb/explore/explore.do?structureId=1T8I (accessed on 23 January 2017).
- Stivers, J.T.; Harris, T.K.; Mildvan, A.S. Vaccinia DNA topoisomerase I: Evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry 1997, 36, 5212–5222. [Google Scholar] [CrossRef] [PubMed]
- Koster, D.A.; Palle, K.; Bot, E.S.M.; Bjornsti, M.-A.; Dekker, N.H. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 2007, 448, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin, A.B.; Stewart, L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 2002, 99, 15387–15392. [Google Scholar] [CrossRef] [PubMed]
- Regairaz, M.; Zhang, Y.-W.; Fu, H.; Agama, K.K.; Tata, N.; Agrawal, S.; Aladjem, M.I.; Pommier, Y. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I–DNA complexes. J. Cell Biol. 2011, 195, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Palle, K.; Vaziri, C. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA Topoisomerase 1 inhibition. Cell Cycle 2011, 10, 1625–1638. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Tuduri, S.; Crabbé, L.; Conti, C.; Tourrière, H.; Holtgreve-Grez, H.; Jauch, A.; Pantesco, V.; De Vos, J.; Thomas, A.; Theillet, C.; et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 2009, 11, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, K.; Mani, C.; Clark, D.W.; Palle, K. Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery. Oncotarget 2016, 7, 12537–12553. [Google Scholar] [PubMed]
- Tsao, Y.P.; D’Arpa, P.; Liu, L.F. The involvement of active DNA synthesis in camptothecin-induced G2 arrest: Altered regulation of p34cdc2/cyclin B. Cancer Res. 1992, 52, 1823–1829. [Google Scholar] [PubMed]
- Kharbanda, S.; Rubin, E.; Gunji, H.; Hinz, H.; Giovanella, B.; Pantazis, P.; Kufe, D. Camptothecin and its derivatives induce expression of the c-jun protooncogene in human myeloid leukemia cells. Cancer Res. 1991, 51, 6636–6642. [Google Scholar] [PubMed]
- Aller, P.; Rius, C.; Mata, F.; Zorrilla, A.; Cabañas, C.; Bellón, T.; Bernabeu, C. Camptothecin induces differentiation and stimulates the expression of differentiation-related genes in U-937 human promonocytic leukemia cells. Cancer Res. 1992, 52, 1245–1251. [Google Scholar] [PubMed]
- Clements, M.K.; Jones, C.B.; Cumming, M.; Daoud, S.S. Antiangiogenic potential of camptothecin and topotecan. Cancer Chemother. Pharmacol. 1999, 44, 411–416. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, J.J.; Shapiro, R.L.; Ren, C.J.; Chuang, N.; Cohen, H.W.; Potmesil, M. Antiangiogenic effects of camptothecin analogues 9-amino-20(S)-camptothecin, topotecan, and CPT-11 studied in the mouse cornea model. Clin. Cancer Res. 1999, 5, 181–187. [Google Scholar] [PubMed]
- Arlt, M.F.; Glover, T.W. Inhibition of topoisomerase I prevents chromosome breakage at common fragile sites. DNA Repair 2010, 9, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, S.B.; Horwitz, M.S. Effects of camptothecin on the breakage and repair of DNA during the cell cycle. Cancer Res. 1973, 33, 2834–2836. [Google Scholar] [PubMed]
- Jayasooriya, R.G.P.T.; Choi, Y.H.; Hyun, J.W.; Kim, G.-Y. Camptothecin sensitizes human hepatoma Hep3B cells to TRAIL-mediated apoptosis via ROS-dependent death receptor 5 upregulation with the involvement of MAPKs. Environ. Toxicol. Pharmacol. 2014, 38, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Strumberg, D.; Pilon, A.A.; Smith, M.; Hickey, R.; Malkas, L.; Pommier, Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5’-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 2000, 20, 3977–3987. [Google Scholar] [CrossRef] [PubMed]
- Priel, E.; Showalter, S.D.; Roberts, M.; Oroszlan, S.; Blair, D.G. The topoisomerase I inhibitor, camptothecin, inhibits equine infectious anemia virus replication in chronically infected CF2Th cells. J. Virol. 1991, 65, 4137–4141. [Google Scholar] [PubMed]
- Bruno, S.; Giaretti, W.; Darzynkiewicz, Z. Effect of camptothecin on mitogenic stimulation of human lymphocytes: Involvement of DNA topoisomerase I in cell transition from G0 to G1 phase of the cell cycle and in DNA replication. J. Cell. Physiol. 1992, 151, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Squires, S.; Ryan, A.J.; Strutt, H.L.; Johnson, R.T. Hypersensitivity of Cockayne’s syndrome cells to camptothecin is associated with the generation of abnormally high levels of double strand breaks in nascent DNA. Cancer Res. 1993, 53, 2012–2019. [Google Scholar] [PubMed]
- Ding, X.; Matsuo, K.; Xu, L.; Yang, J.; Zheng, L. Optimized combinations of bortezomib, camptothecin, and doxorubicin show increased efficacy and reduced toxicity in treating oral cancer. Anticancer Drugs 2015, 26, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Walter, J.C. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair 2014, 19, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Ray Chaudhuri, A.; Hashimoto, Y.; Herrador, R.; Neelsen, K.J.; Fachinetti, D.; Bermejo, R.; Cocito, A.; Costanzo, V.; Lopes, M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012, 19, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/c9911pis.pdf (accessed on 23 November 2016).
- Jaxel, C.; Kohn, K.W.; Wani, M.C.; Wall, M.E.; Pommier, Y. Structure-activity study of the actions of camptothecin derivatives on mammalian topoisomerase I: Evidence for a specific receptor site and a relation to antitumor activity. Cancer Res. 1989, 49, 1465–1469. [Google Scholar] [PubMed]
- Takagi, K.; Dexheimer, T.S.; Redon, C.; Sordet, O.; Agama, K.; Lavielle, G.; Pierré, A.; Bates, S.E.; Pommier, Y. Novel E-ring camptothecin keto analogues (S38809 and S39625) are stable, potent, and selective topoisomerase I inhibitors without being substrates of drug efflux transporters. Mol. Cancer Ther. 2007, 6, 3229–3238. [Google Scholar] [CrossRef] [PubMed]
- Hande, K.R. Etoposide: Four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 1998, 34, 1514–1521. [Google Scholar] [CrossRef]
- Available online: http://www.rcsb.org/pdb/explore/explore.do?structureId=3QX3 (accessed on 23 January 2017).
- Liu, L.F.; Rowe, T.C.; Yang, L.; Tewey, K.M.; Chen, G.L. Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 1983, 258, 15365–15370. [Google Scholar] [PubMed]
- Gibson, E.G.; King, M.M.; Mercer, S.L.; Deweese, J.E. Two-Mechanism Model for the Interaction of Etoposide Quinone with Topoisomerase IIα. Chem. Res. Toxicol. 2016, 29, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Li, T.-K.; Farh, L.; Lin, L.-Y.; Lin, T.-S.; Yu, Y.-J.; Yen, T.-J.; Chiang, C.-W.; Chan, N.-L. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 2011, 333, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.P.; Jablonksy, M.J.; Shadid, M.; Romaine, I.; Dunlap, N.; Anklin, C.; Graves, D.E.; Osheroff, N. Substituents on etoposide that interact with human topoisomerase IIalpha in the binary enzyme-drug complex: Contributions to etoposide binding and activity. Biochemistry 2008, 47, 4501–4509. [Google Scholar] [CrossRef] [PubMed]
- Wilstermann, A.M.; Bender, R.P.; Godfrey, M.; Choi, S.; Anklin, C.; Berkowitz, D.B.; Osheroff, N.; Graves, D.E. Topoisomerase II—Drug interaction domains: Identification of substituents on etoposide that interact with the enzyme. Biochemistry 2007, 46, 8217–8225. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.A.; Mercer, S.L.; Osheroff, N.; Deweese, J.E. Etoposide quinone is a redox-dependent topoisomerase II poison. Biochemistry 2011, 50, 5660–5667. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, M.; Shinohara, A.; Shinohara, M. Canonical non-homologous end joining in mitosis induces genome instability and is suppressed by M-phase-specific phosphorylation of XRCC4. PLoS Genet. 2014, 10, e1004563. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Rybak, P.; Dobrucki, J.; Traganos, F.; Darzynkiewicz, Z. Relationship of DNA damage signaling to DNA replication following treatment with DNA topoisomerase inhibitors camptothecin/topotecan, mitoxantrone, or etoposide. Cytometry A 2012, 81, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Montecucco, A.; Rossi, R.; Ferrari, G.; Scovassi, A.I.; Prosperi, E.; Biamonti, G. Etoposide Induces the Dispersal of DNA Ligase I from Replication Factories. Mol. Biol. Cell 2001, 12, 2109–2118. [Google Scholar] [CrossRef] [PubMed]
- Holm, C.; Covey, J.M.; Kerrigan, D.; Pommier, Y. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 1989, 49, 6365–6368. [Google Scholar] [PubMed]
- Austin, C.A.; Sng, J.H.; Patel, S.; Fisher, L.M. Novel HeLa topoisomerase II is the II beta isoform: Complete coding sequence and homology with other type II topoisomerases. Biochim. Biophys. Acta 1993, 1172, 283–291. [Google Scholar] [CrossRef]
- Niimi, A.; Suka, N.; Harata, M.; Kikuchi, A.; Mizuno, S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma 2001, 110, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.-G.; Lunyak, V.V.; Perissi, V.; Garcia-Bassets, I.; Rose, D.W.; Glass, C.K.; Rosenfeld, M.G. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 2006, 312, 1798–1802. [Google Scholar] [CrossRef] [PubMed]
- Azarova, A.M.; Lyu, Y.L.; Lin, C.-P.; Tsai, Y.-C.; Lau, J.Y.-N.; Wang, J.C.; Liu, L.F. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. USA 2007, 104, 11014–11019. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Bromke, A.; Bryant, D.W.; Gupta, R.; Singh, B.; McCalla, D.R. Etoposide (VP16) and teniposide (VM26): Novel anticancer drugs, strongly mutagenic in mammalian but not prokaryotic test systems. Mutagenesis 1987, 2, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Muslimović, A.; Nyström, S.; Gao, Y.; Hammarsten, O. Numerical Analysis of Etoposide Induced DNA Breaks. PLoS ONE 2009, 4, e5859. [Google Scholar] [CrossRef]
- Álvarez-Quilón, A.; Serrano-Benítez, A.; Lieberman, J.A.; Quintero, C.; Sánchez-Gutiérrez, D.; Escudero, L.M.; Cortés-Ledesma, F. ATM specifically mediates repair of double-strand breaks with blocked DNA ends. Nat. Commun. 2014, 5, 3347. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Nakano, M.; Nakashima, A.; Onishi, K.; Yamao, S.; Enari, M.; Kikkawa, U.; Kamada, S. Identification of cellular senescence-specific genes by comparative transcriptomics. Sci. Rep. 2016, 6, 31758. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Alsop, A.E.; Noori, T.; Lufti, M.; Iyer, S.; Simpson, K.J.; Bird, P.I.; Kluck, R.M.; Johnstone, R.W.; Trapani, J.A. Epigenetic control of mitochondrial cell death through PACS1-mediated regulation of BAX/BAK oligomerization. Cell Death Differ. 2017. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Desponds, C.; Eren, R.O.; Quadroni, M.; Thome, M.; Fasel, N. Caspase-mediated cleavage of raptor participates in the inactivation of mTORC1 during cell death. Cell Death Discov. 2016, 2, 16024. [Google Scholar] [CrossRef] [PubMed]
- Brekman, A.; Singh, K.E.; Polotskaia, A.; Kundu, N.; Bargonetti, J. A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res. 2011, 13, R3. [Google Scholar] [CrossRef] [PubMed]
- Soubeyrand, S.; Pope, L.; Haché, R.J.G. Topoisomerase IIα-dependent induction of a persistent DNA damage response in response to transient etoposide exposure. Mol. Oncol. 2010, 4, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Velma, V.; Carrero, Z.I.; Allen, C.B.; Hebert, M.D. Coilin levels modulate cell cycle progression and γH2AX levels in etoposide treated U2OS cells. FEBS Lett. 2012, 586, 3404–3409. [Google Scholar] [CrossRef] [PubMed]
- Dehennaut, V.; Loison, I.; Dubuissez, M.; Nassour, J.; Abbadie, C.; Leprince, D. DNA double-strand breaks lead to activation of hypermethylated in cancer 1 (HIC1) by SUMOylation to regulate DNA repair. J. Biol. Chem. 2013, 288, 10254–10264. [Google Scholar] [CrossRef] [PubMed]
- Paget, S.; Dubuissez, M.; Dehennaut, V.; Nassour, J.; Harmon, B.T.; Spruyt, N.; Loison, I.; Abbadie, C.; Rood, B.R.; Leprince, D. HIC1 (hypermethylated in cancer 1) SUMOylation is dispensable for DNA repair but is essential for the apoptotic DNA damage response (DDR) to irreparable DNA double-strand breaks (DSBs). Oncotarget 2017, 8, 2916–2935. [Google Scholar] [CrossRef] [PubMed]
- Sypniewski, D.; Bednarek, I.; Gałka, S.; Loch, T.; Błaszczyk, D.; Sołtysik, D. Cytotoxicity of etoposide in cancer cell lines in vitro after BCL-2 and C-RAF gene silencing with antisense oligonucleotides. Acta Pol. Pharm. 2013, 70, 87–97. [Google Scholar]
- Rybak, P.; Hoang, A.; Bujnowicz, L.; Bernas, T.; Berniak, K.; ZarÄ Bski, M.A.; Darzynkiewicz, Z.; Dobrucki, J. Low level phosphorylation of histone H2AX on serine 139 (γH2AX) is not associated with DNA double-strand breaks. Oncotarget 2016, 7, 49574–49587. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cui, H.; Fang, J.; Deng, H.; Kuang, P.; Guo, H.; Wang, X.; Zhao, L. Glutamine deprivation plus BPTES alters etoposide- and cisplatin-induced apoptosis in triple negative breast cancer cells. Oncotarget 2016, 7, 54691–54701. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lopez, A.M.; Xenaki, D.; Eden, T.O.; Hickman, J.A.; Chresta, C.M. MDM2 mediated nuclear exclusion of p53 attenuates etoposide-induced apoptosis in neuroblastoma cells. Mol. Pharmacol. 2001, 59, 135–143. [Google Scholar]
- Litwiniec, A.; Gackowska, L.; Helmin-Basa, A.; Żuryń, A.; Grzanka, A. Low-dose etoposide-treatment induces endoreplication and cell death accompanied by cytoskeletal alterations in A549 cells: Does the response involve senescence? The possible role of vimentin. Cancer Cell Int. 2013, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Talegaonkar, S.; Khar, R.K.; Jaggi, M. A validated stability-indicating LC method for estimation of etoposide in bulk and optimized self-nano emulsifying formulation: Kinetics and stability effects. Saudi Pharm. J. 2013, 21, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Product_Information_Sheet/e1383pis.pdf (accessed on 23 Novermber 2016).
- Wrasidlo, W.; Schröder, U.; Bernt, K.; Hübener, N.; Shabat, D.; Gaedicke, G.; Lode, H. Synthesis, hydrolytic activation and cytotoxicity of etoposide prodrugs. Bioorg Med Chem Lett. 2002, 12, 557–560. [Google Scholar] [CrossRef]
- Jokić, M.; Vlašić, I.; Rinneburger, M.; Klümper, N.; Spiro, J.; Vogel, W.; Offermann, A.; Kümpers, C.; Fritz, C.; Schmitt, A.; et al. Ercc1 Deficiency Promotes Tumorigenesis and Increases Cisplatin Sensitivity in a Tp53 Context-Specific Manner. Mol. Cancer Res. 2016, 14, 1110–1123. [Google Scholar] [CrossRef] [PubMed]
- Felix, C.A.; Walker, A.H.; Lange, B.J.; Williams, T.M.; Winick, N.J.; Cheung, N.K.; Lovett, B.D.; Nowell, P.C.; Blair, I.A.; Rebbeck, T.R. Association of CYP3A4 genotype with treatment-related leukemia. Proc. Natl. Acad. Sci. USA 1998, 95, 13176–13181. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J.G.; Edick, M.J.; Relling, M.V. Etoposide induces chimeric Mll gene fusions. FASEB J. 2004, 18, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Thirman, M.J.; Gill, H.J.; Burnett, R.C.; Mbangkollo, D.; McCabe, N.R.; Kobayashi, H.; Ziemin-van der Poel, S.; Kaneko, Y.; Morgan, R.; Sandberg, A.A. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N. Engl. J. Med. 1993, 329, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Cerveira, N.; Lisboa, S.; Correia, C.; Bizarro, S.; Santos, J.; Torres, L.; Vieira, J.; Barros-Silva, J.D.; Pereira, D.; Moreira, C.; et al. Genetic and clinical characterization of 45 acute leukemia patients with MLL gene rearrangements from a single institution. Mol. Oncol. 2012, 6, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, F.; Zhang, Z.; Chen, Y.; Lin, Y.; Wang, J. Design, synthesis and evaluation of the multidrug resistance-reversing activity of pyridine acid esters of podophyllotoxin in human leukemia cells. Bioorg. Med. Chem. Lett. 2016, 26, 4466–4471. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-I.; Su, C.-C.; Yang, C.-Y.; Hung, D.-Z.; Lin, C.-T.; Lu, T.-H.; Liu, S.-H.; Huang, C.-F. Etoposide induces pancreatic β-cells cytotoxicity via the JNK/ERK/GSK-3 signaling-mediated mitochondria-dependent apoptosis pathway. Toxicol. Vitro 2016, 36, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.G.; Morales, C.C.; Wallace, T.C.; Plotkin, L.I.; Bellido, T. Avenanthramides Prevent Osteoblast and Osteocyte Apoptosis and Induce Osteoclast Apoptosis in Vitro in an Nrf2-Independent Manner. Nutrients 2016, 8, 423. [Google Scholar] [CrossRef] [PubMed]
- Papież, M.A.; Krzyściak, W.; Szade, K.; Bukowska-Straková, K.; Kozakowska, M.; Hajduk, K.; Bystrowska, B.; Dulak, J.; Jozkowicz, A. Curcumin enhances the cytogenotoxic effect of etoposide in leukemia cells through induction of reactive oxygen species. Drug Des. Dev. Ther. 2016, 10, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lu, C.; Zhang, X.; Li, J.; Jiang, H. Targeted delivery of etoposide to cancer cells by folate-modified nanostructured lipid drug delivery system. Drug Deliv. 2016, 23, 1838–1845. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, G.S.; Wallace, H.M. Changes in polyamine catabolism in HL-60 human promyelogenous leukaemic cells in response to etoposide-induced apoptosis. Biochem. J. 1999, 337 Pt 1, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ehrenshaft, M.; Tokar, E.J.; Mason, R.P.; Sinha, B.K. Nitric oxide inhibits topoisomerase II activity and induces resistance to topoisomerase II-poisons in human tumor cells. Biochim. Biophys. Acta 2016, 1860, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Lyu, Y.L.; Lin, C.-P.; Zhou, N.; Azarova, A.M.; Wood, L.M.; Liu, L.F. A protease pathway for the repair of topoisomerase II–DNA covalent complexes. J. Biol. Chem. 2006, 281, 35997–36003. [Google Scholar] [CrossRef] [PubMed]
- Ledesma, F.C.; El Khamisy, S.F.; Zuma, M.C.; Osborn, K.; Caldecott, K.W. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 2009, 461, 674–678. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, T.; Baer, R.; Gottesman, M.; Gautier, J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II–DNA adducts. J. Cell Biol. 2016, 212, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Quennet, V.; Beucher, A.; Barton, O.; Takeda, S.; Löbrich, M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 2011, 39, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- Adachi, N.; Suzuki, H.; Iiizumi, S.; Koyama, H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: Implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 2003, 278, 35897–35902. [Google Scholar] [CrossRef] [PubMed]


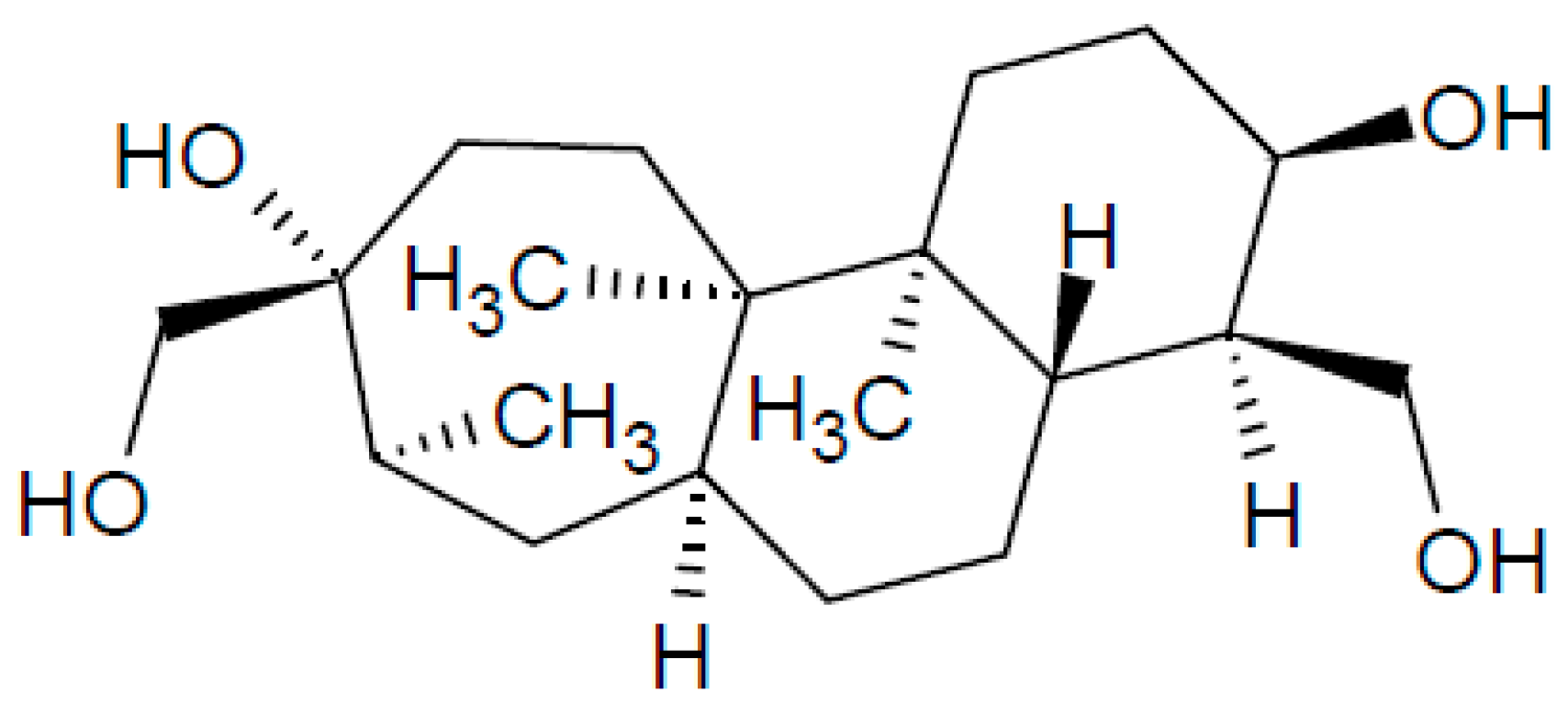
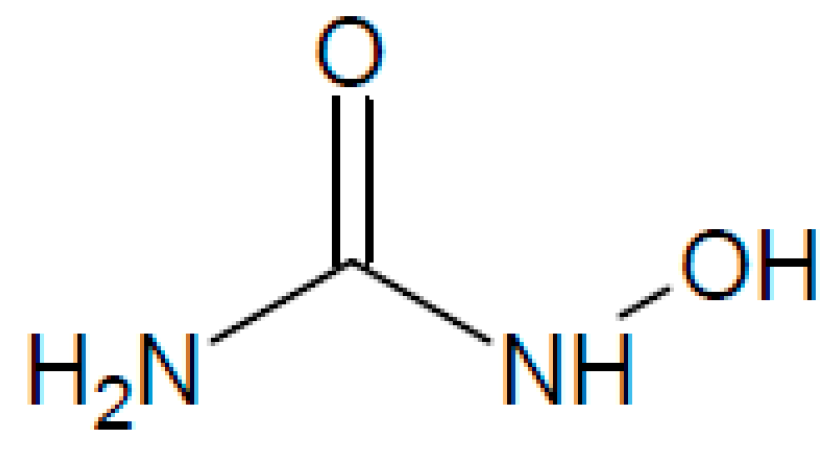
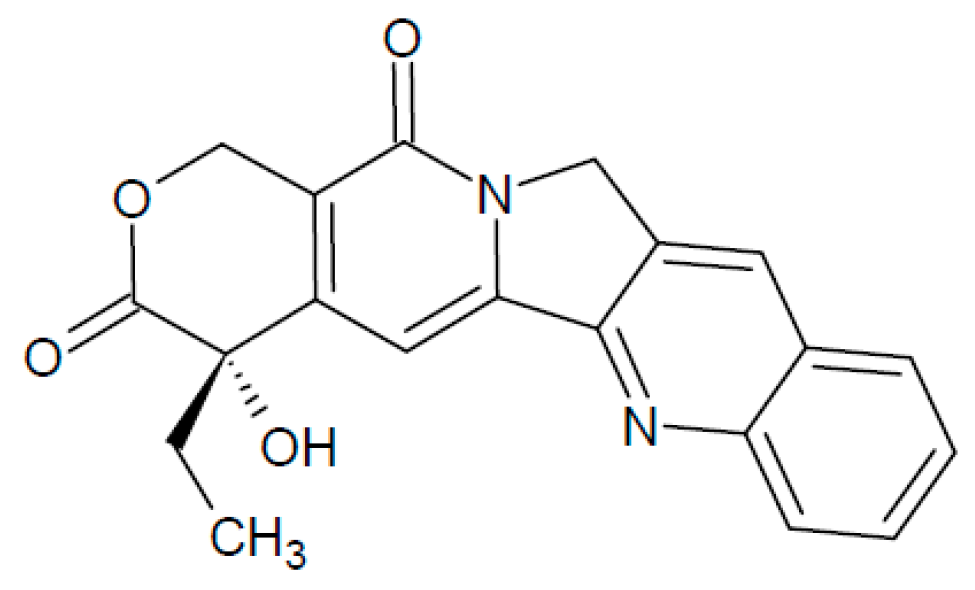
| Concentration | Incubation Time | Observed Effect | Cell Line | Reference |
|---|---|---|---|---|
| 300 μM | 2 h | increase in polyADP ribosylation | O-342 rat ovarian tumour cells | [137] |
| 100 μM | 2 h before IR | sensitization to γ-radiation | hypoxic V-79 Chinese hamster cells | [138] |
| 100 μM | 2 h | increase in polyADP ribosylation | CV-l monkey cells | [139] |
| <20 μg/mL (<66 μM) | 5 h | block of rRNA synthesis block of DNA replication | Hela | [140] |
| 15 μM | 1 h | induction of SCE (sister chromatid exchange) decreased cell survival | 6 primary human tumour cell culture | [141] |
| 10–30 μM | 24 h, 48 h | induction of apoptosis | 224 (melanoma cells) HCT116 | [142] |
| 10 μM | 24 h | increase in antiapoptotic Bcl-2 mRNA synthesis (regulated by PKC and Akt2) | KLE HEC-1-A Ishikawa MCF-7 | [143] |
| 2–10 μM | 72 h | induction of apoptosis | 224 (melanoma cells) HCT116 | [142] |
| 5 μM | 24 h | increase in p53 stability activation of ATR increased p53(ser15) phosphorylation | A2780 | [144] |
| 5 μM | 24 h | activation of p21 activation of CHK2 increased p53(ser20) phosphorylation | HCT116 | [144] |
| 5 μM | 24 h | induction of mitochondrial reactive oxygen species (ROS) response | A549 PC3 MEF | [143] |
| 2 μM | 24 h | G2/M arrest, subapoptic damage | MSC | [145] |
| >2 μM | 24 h | decreased proliferation rate induction of apoptosis | TGCT H12.1 TGCT 2102EP | [145] |
| 1–4 μg/mL | 2 h | block of DNA synthesis | L1210/0 cells | [146] |
| block of transcription | ||||
| G2 arrest | ||||
| apoptosis | ||||
| 2 μg/mL | 48 h 144 h, 168 h | inhibition of mtDNA replication inhibition of mitochondrial genes transcription | Dorsal root ganglion (DRG) sensory neurons | [147] |
| 1 μg/mL | 2 h | transient G2 arrest | Hela | [148] |
| 3.0 μM | 4 h before | block of NHEJ | A2780 | [138] |
| 0.2–0.8 μM | IR 0.5 Gy | cisPt-IR synergistic interaction | MO59J MO59K | [138] |
| 4 h | ||||
| 1–2.5 μM | 24 h–48 h | block of DNA replication followed by cell apoptosis | Hela | [149] |
| 0.3–1 μM | overnight | inhibition of RNA polymerase II-dependent transcription | Hela XPF | [144] |
| 0.6 μM | 2 h | 90% reduction in clonogenic capacity detected after 7 days CHK1 phosphorylation causing CHK1 dependent S phase arrest | Hela | [148] |
| 0.5 μM | 24 h 48 h | loss of telomeres (TEL), or TEL repeats cell death | Hela | [139] |
| Concentration | Incubation Time | Observed Effect | Cell Line | Reference |
|---|---|---|---|---|
| 0.2 mM | 16 h, 10 h | formation of anaphase bridges and micronuclei | HeLa | [204] |
| 30 μM | 6 h | stalled replication forks | HCT116 | [205] |
| 30 μM | 6 h | stalled replication forks | PD20 cells Bloom syndrome cells | [206] |
| 5 μg/mL (14.3 μM) | 4 h | DNA repair synthesis inhibition sensitization towards TNF treatment | L929 ovarian cancer cells A2780 | [202] |
| 5 μg/mL (14.3 μM) | 2–8 h | S phase arrest kinetics and mechanism study | RKO 293T MEF | [207] |
| 2.5 μg/mL (7.15 μM) | 1 h | inhibition of DNA synthesis and DNA repair | Normal and XPA deficient human fibroblasts | [203] |
| 10 μM | 15 h | cell cycle synchronisation at the G1/S boundary | REF-52 HeLa | [208] |
| 5–25 μM | 24 h | inhibition of replicative polymerases | Werner syndrome cells Bloom syndrome cells | [209,210] |
| 1 μM | 1–24 h | CFS induction | HEK293T | [210] |
| 1 μM | 24 h | CFS induction | MEF HeLa | [211] |
| 0.5 μM | 2 h | transient attenuation of DNA synthesis, | DT40 | [212] |
| 0.1 μM | 24 h | study of chromosome integrity and replication | ||
| 0.4 μM | 24 h | CFS induction | U-2 OS | [213] |
| 0.1 μM 0.2 μM | 16 h | replication stress observed on telomeres | hESC (UCSF4) | [214] |
| 0.2 μM | 2 weeks | irreversible senescence induction | REF-52 | [74] |
| 0.2 μM | 24 h | CFS induction | BJ-hTERT | [215] |
| 0.05 μM 0.4 μM | 24 h | CFS induction | Werner syndrome fibroblasts AG11395 cells | [216] |
| 0.3 μM | 48 h | increased incidence of mitotic extra chromosomes replication stress | V79 hamster cell lines | [217] |
| 0.3 μM | 72 h | replication stress | Human fibroblasts HGMDFN090 | [199] |
| 2 μg/mL | not indicated | replication block | BJ BJ-tert HMEC | [197] |
| 0.2 μM | 7–24 h | cell synchronization | HeLa | [184] |
| Concentration | Incubation Time | Effect | Cell Line | Reference |
|---|---|---|---|---|
| 200 mM | 2 h | replication block | yeast cells | [240] |
| 10–200 mM | 3 h | replication block replication fork (RF) restart | yeast cells | [241] |
| 5 mM | 1 h | replication block | HEK293 | [242] |
| 2 mM | 3 h | replication block | ||
| 50 μM–5 mM | 40 min–2 h | replication stress | 293T mouse ES cells | [243] |
| 2 mM | 1 h, 24 h | replication stress replication block | HCC1937 MCF7 | [244] |
| 2 mM | 16 h | replication block | HEK293 | [245] |
| 2 mM | 24 h | DNA damage induction during S phase | U-2 OS 293T | [246] |
| 2 mM | 15 h | replication block cell cycle synchronisation at the G1/S boundary | REF-52 HeLa | [208] |
| 2 mM | 5 h | dNTP depletion | REF52 | [74] |
| 2 mM | 3 h | chromosomal aberrations FANCD2 pathway involvement | lymphoblastoid cell lines | [247] |
| 1 mM | overnight | replication block | MCF7 | [248] |
| 0.5 mM | 5 h–10 h | replication block | U-2OS | [249] |
| 2 mM | 2 h–24 h | replication block | ||
| 0.5 mM | 90 min | nucleotides depletion stalled RF w/o DSBs formation | MEF | [250] |
| 0.1–0.5 mM | 2 h–72 h | γ-globin gene expression | K562 | [251] |
| 0.1–0.5 mM | 2 h–8 h | replication stress | PC3 | [252] |
| 0.2–0.4 mM | 4 days | cell differentiation ERK signalling pathway inhibition p38 signal transduction activation | K562 | [253] |
| 0.3 mM | 10 days | microsatellite instability upon FANCJ depletion | GM08402 HeLa PD20F | [254] |
| 0.15–0.2 mM | 2 weeks | irreversible senescence induction | REF-52 | [74] |
| 0.2 mM | 2 h–7 h | replication stress | MEF | [255] |
| 0.15 mM | 2 h | p53 activation | REF52 | [74] |
| 50–200 μM | 20 h | HIF1 induction eNOS induction | HUVEC | [256] |
| 25–200 μM | 72 h | induction of apoptosis | AML cell lines (MV4-11, OCI-AML3, MOLM-13, and HL-60) | [257] |
| 5 μM–0.5 mM | 48 h | replication stress | V79 hamster cells | [217] |
| 2 μM | 12 h | replication stress | H1299 | [258] |
| Concentration | Incubation Time | Observed Effect | Cell Line | Reference |
|---|---|---|---|---|
| 20 μM | 30 min | DNA fragmentation in G1 and S phase cells | Hela | [292] |
| 10 μM | 24 h | increase in cell sensitivity to TRAIL-mediated apoptosis | Hep3B | [293] |
| 10 μM | 4 h | formation of replication mediated DNA DSBs | HT29 | [294] |
| 5 μM | 60 min | inhibition of RNA synthesis | CSA | [295] |
| 1 μM | 60 min | inhibition of DNA synthesis | CSB | [296] |
| 1 μM | 60 min | replication block DSB formation cell death | U2OS | [297] |
| 1 μM | 60 min | formation of stabilised TopoI-cc complex DSB formation phosphorylation of CHK1 (S317) CHK2 (T68), RPA (S4/S8) | HCT116 | [294] |
| 1 μM | 60 min | inhibition of DNA replication suggested DNA DSB formation | L1210 mouse lymphoblastic leukaemic cells | [293] |
| 200 nM–1 μM | 50 min | DSB formation | CSB | [298] |
| 100 nM–10 nM | 60 min | DSB formation | HCT116 | [299] |
| 25 nM | 60 min | checkpoint activation (ATM-CHK2, ATR-CHK1) replication fork stalling replication fork reversal formation of specific DNA structures | U-2O-S | [300] |
| 10 nM–100 nM | 60 min | inhibition of EIAV (equine infectious anemia virus) replication | CF2Th | [295] |
| 10 nM–20 nM | 60 min | inhibition of HIV-1 replication block of viral protein expression | H9 | [281] |
| 6 nM | 6 h | accumulation of cells in early S phase | Normal lymphocytes | [296] |
| 24 h | apoptosis, DNA fragmentation | MOLT-4 | ||
| 6.25 nM | 48 h | specific suppression of oral cancer cells growth | KB oral cancer cells | [281] |
| 2.5 nM | 48 h | increase in SCE upon depletion of Fbh1 helicase | BJ | [281] |
| Concentration | Incubation Time | Effect | Cell Line | References |
|---|---|---|---|---|
| up to 450 µM | 40 min | SSB and DSB formation, induction of H2AX phosphorylation with slow kinetics | SV-40 transformed human fibroblasts G361 | [323] |
| 1–100 µM | 30 min | formation of TopoII-blocked DSBs, activation of ATM-mediated repair | MEF HEK293T BJ1 AT | [324] |
| 2–100 μM | 6 h–48 h | senescence, apoptosis induction of p53 response | HepG2 U2OS | [325] |
| 2–100 μM | 1–3 h | disassembly of replication factories | AT1 BR AT3 BR HeLa | [315] |
| 50–100 μM | 3–6 h/16 h | apoptosis (activation of intrinsic (mitochondrial) pathway) | Hela HCT116 | [326] |
| 50 μM | 15 h | apoptosis | BJAB Hut78 | [327] |
| 50 μM | 48 h | growth arrest (accumulation of cells at G2/M boundary) induction of p53 response | MCF-7 ZR75-1 T-47D | [328] |
| 25 µM | 1 h | SSB and DSB formation γH2AX, pATM, pDNA-PKcs, MDC1 foci formation persisting DSBs cell death | HeLa HCT116 | [329] |
| 20 μM | 16 h | increase in γH2AX levels reduction of proliferation rate (accumulation of cells in S and G2/M boundary) | U2OS | [330] |
| 20 μM | 1 h | repairable DSBs | HEK293T COS-7 BJ-hTERT H1299 | [331,332] |
| 16 h | irrepairable DSBs, ATM-dependent HIC1 SUMOylation, induction of p53-dependent apoptotic response | |||
| 20 µM | 1–5 h | apoptosis | A549 HeLa, T24 | [333] |
| 10 μM | 1 h | DNA damage induction | A549 | [334] |
| 1–10 μM | 48 h | apoptosis | HCC1937 BT-549 | [335] |
| 8 μM | 1 h | induction of p53 response, | SH-SY-5Y SH-EP1 | [336] |
| 0.75–3 µM | 72 h | senescence, apoptosis | A549 | [337] |
| 0.75 μM | 24 h | cell cycle arrest in G2/M phase, DNA damage induction, induction of p53 response | MSC TGCT H12.1 TGCT 2102EP | [145] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vesela, E.; Chroma, K.; Turi, Z.; Mistrik, M. Common Chemical Inductors of Replication Stress: Focus on Cell‐Based Studies. Biomolecules 2017, 7, 19. https://doi.org/10.3390/biom7010019
Vesela E, Chroma K, Turi Z, Mistrik M. Common Chemical Inductors of Replication Stress: Focus on Cell‐Based Studies. Biomolecules. 2017; 7(1):19. https://doi.org/10.3390/biom7010019
Chicago/Turabian StyleVesela, Eva, Katarina Chroma, Zsofia Turi, and Martin Mistrik. 2017. "Common Chemical Inductors of Replication Stress: Focus on Cell‐Based Studies" Biomolecules 7, no. 1: 19. https://doi.org/10.3390/biom7010019





