Modulation of the Proteostasis Machinery to Overcome Stress Caused by Diminished Levels of t6A‐Modified tRNAs in Drosophila
Abstract
:1. Introduction
2. Results
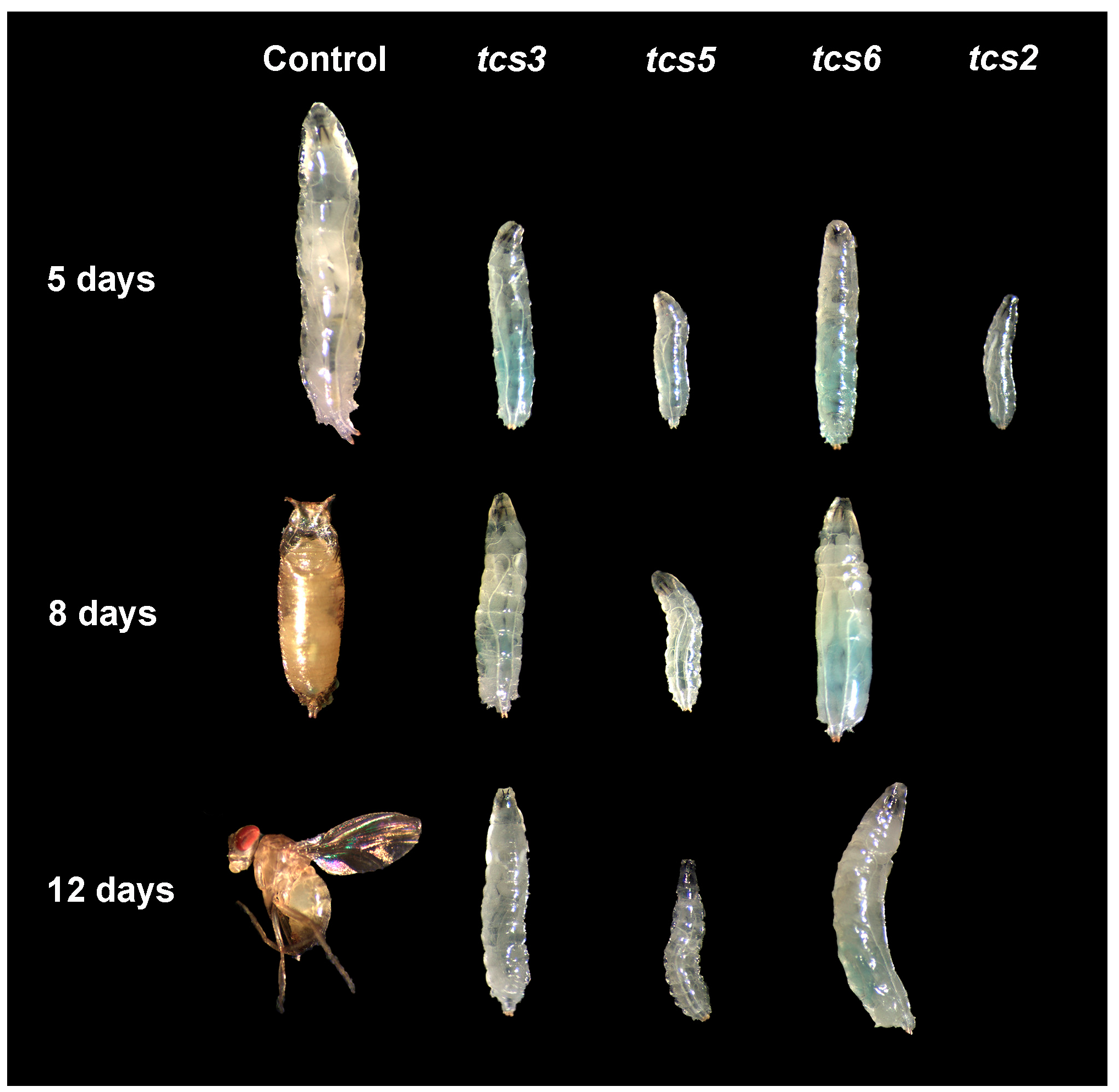
2.1. t6A Synthetic Machinery is Required for Larval Growth
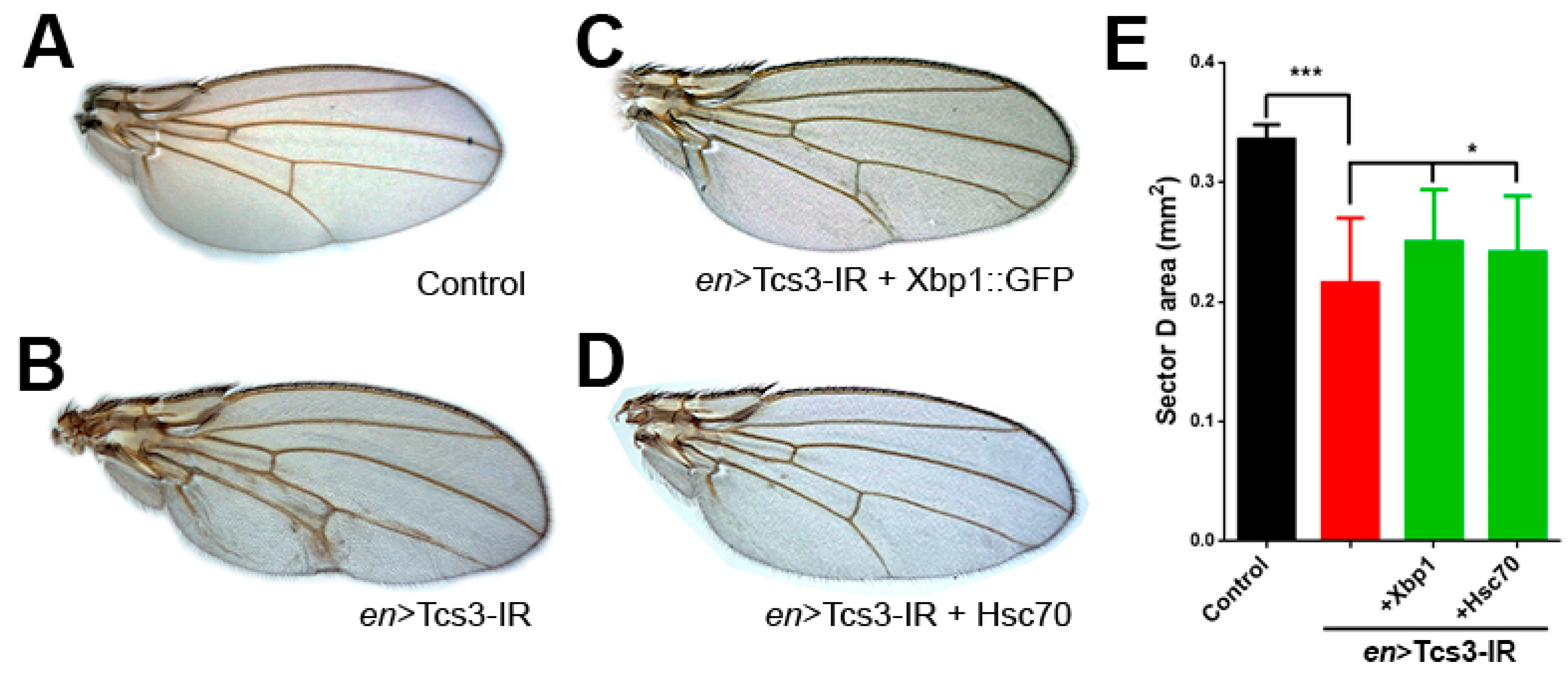
2.2. Tcs3 is Required for Imaginal Cell Survival
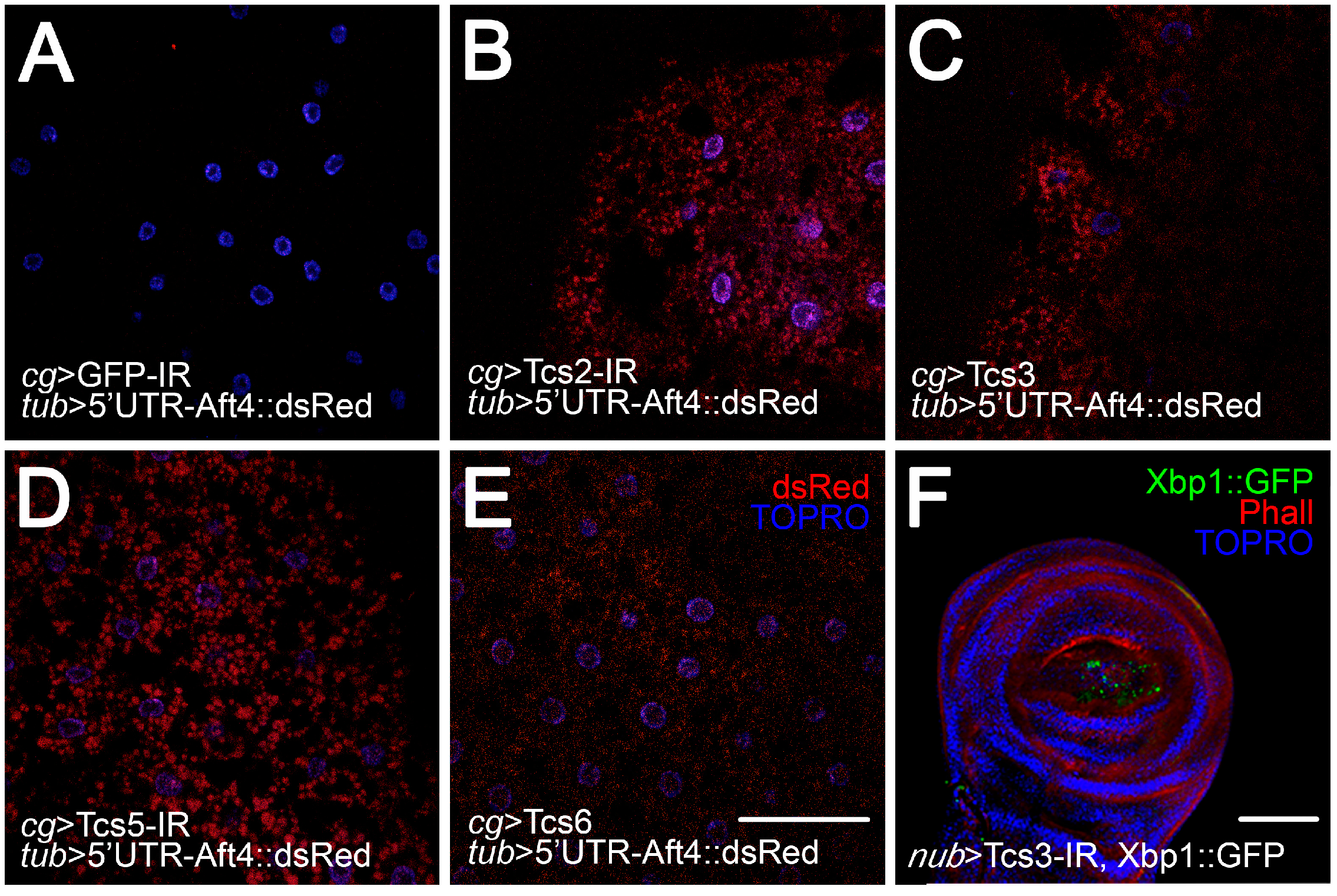
2.3. Activation of UPR upon Silencing of t6A Synthetic Machinery
3. Discussion
4. Materials and Methods
4.1. Fly Husbandry and Fly Stocks
4.2. Morphological, Morphometric and Statistical Analysis
4.3. Immunofluorescence
4.4. Flow Cytometry
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer rnas. Ann. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.M.; Pavon-Eternod, M.; Pan, T.; de Pouplana, L.R. A role for tRNA modifications in genome structure and codon usage. Cell 2012, 149, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Guo, J.; Cha, J.; Chae, M.; Chen, S.; Barral, J.M.; Sachs, M.S.; Liu, Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 2013, 495, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Hori, H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front. Genet. 2014, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Daugeron, M.C.; Lenstra, T.L.; Frizzarin, M.; El Yacoubi, B.; Liu, X.; Baudin-Baillieu, A.; Lijnzaad, P.; Decourty, L.; Saveanu, C.; Jacquier, A.; et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011, 39, 6148–6160. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Hatin, I.; Deutsch, C.; Kahveci, T.; Rousset, J.P.; Iwata-Reuyl, D.; Murzin, A.G.; de Crecy-Lagard, V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011, 30, 882–893. [Google Scholar] [CrossRef] [PubMed]
- Perrochia, L.; Crozat, E.; Hecker, A.; Zhang, W.; Bareille, J.; Collinet, B.; van Tilbeurgh, H.; Forterre, P.; Basta, T. In vitro biosynthesis of a universal t6A tRNA modification in archaea and eukarya. Nucleic Acids Res. 2013, 41, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, C.; El Yacoubi, B.; de Crecy-Lagard, V.; Iwata-Reuyl, D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012, 287, 13666–13673. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Benitez, D.; Thiaville, P.C.; de Crecy-Lagard, V.; Glavic, A. The levels of a universally conserved trna modification regulate cell growth. J. Biol. Chem. 2015, 290, 18699–18707. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Smibert, P.; Zhao, X.; Hu, J.F.; Ramroop, J.; Kellner, S.M.; Benton, M.A.; Govind, S.; Dedon, P.C.; Sternglanz, R.; et al. An extensive allelic series of drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA 2015, 21, 2103–2118. [Google Scholar] [CrossRef] [PubMed]
- Elkins, B.N.; Keller, E.B. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry 1974, 13, 4622–4628. [Google Scholar] [CrossRef] [PubMed]
- Korner, A.; Soll, D. N-(purin-6-ylcarbamoyl)threonine: Biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett. 1974, 39, 301–306. [Google Scholar] [CrossRef]
- Chheda, G.B.; Hong, C.I.; Piskorz, C.F.; Harmon, G.A. Biosynthesis of N-(purin-6-ylcarbamoyl)-l-threonine riboside. Incorporation of l-threonine in vivo into modified nucleoside of transfer ribonucleic acid. Biochem. J. 1972, 127, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Perrochia, L.; Guetta, D.; Hecker, A.; Forterre, P.; Basta, T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A trna modification. Nucleic Acids Res. 2013, 41, 9484–9499. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol. 2014, 11, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Ibar, C.; Cataldo, V.F.; Vasquez-Doorman, C.; Olguin, P.; Glavic, A. Drosophila p53-related protein kinase is required for PI3K/TOR pathway-dependent growth. Development 2013, 140, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Legendre, R.; Rojas-Benitez, D.; Baudin-Baillieu, A.; Hatin, I.; Chalancon, G.; Glavic, A.; Namy, O.; de Crecy-Lagard, V. Global translational impacts of the loss of the trna modification t6A in yeast. Microb. Cell 2016, 3, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Benitez, D.; Ibar, C.; Glavic, A. The drosophila EKC/KEOPS complex: Roles in protein synthesis homeostasis and animal growth. Fly 2013, 7, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C.; Chevet, E.; Oakes, S.A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 2015, 17, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [PubMed]
- Kim, K.; Lee, Y.S.; Harris, D.; Nakahara, K.; Carthew, R.W. The RNAi pathway initiated by Dicer-2 in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Perkins, L.A.; Holderbaum, L.; Tao, R.; Hu, Y.; Sopko, R.; McCall, K.; Yang-Zhou, D.; Flockhart, I.; Binari, R.; Shim, H.S.; et al. The transgenic RNAi project at harvard medical school: Resources and validation. Genetics 2015, 201, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Comjean, A.; Roesel, C.; Vinayagam, A.; Flockhart, I.; Zirin, J.; Perkins, L.; Perrimon, N.; Mohr, S.E. FlyRNAi.Org—The database of the drosophila RNAi screening center and transgenic RNAi project: 2017 update. Nucleic Acids Res. 2017, 45, D672–D678. [Google Scholar] [CrossRef] [PubMed]
- Morata, G. How drosophila appendages develop. Nat. Rev. Mol. Cell Biol. 2001, 2, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Brower, D.L. Engrailed gene expression in Drosophila imaginal discs. EMBO J. 1986, 5, 2649–2656. [Google Scholar] [PubMed]
- Vermes, I.; Haanen, C.; Reutelingsperger, C. Flow cytometry of apoptotic cell death. J. Immunol. Methods 2000, 243, 167–190. [Google Scholar] [CrossRef]
- Ohtsubo, T.; Kamada, S.; Tsujimoto, Y. Inhibition of apoptosis by a baculovirus p35 gene. Nihon Rinsho. J. J. Clin. Med. 1996, 54, 1907–1911. [Google Scholar]
- Hay, B.A.; Wolff, T.; Rubin, G.M. Expression of baculovirus p35 prevents cell death in Drosophila. Development 1994, 120, 2121–2129. [Google Scholar] [PubMed]
- Igaki, T.; Kanda, H.; Yamamoto-Goto, Y.; Kanuka, H.; Kuranaga, E.; Aigaki, T.; Miura, M. Eiger, a TNF superfamily ligand that triggers the drosophila JNK pathway. EMBO J. 2002, 21, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C. The biological meaning of the UPR. Nat. Rev. Mol. Cell Biol. 2013, 14, 404. [Google Scholar] [CrossRef] [PubMed]
- Demay, Y.; Perochon, J.; Szuplewski, S.; Mignotte, B.; Gaumer, S. The PERK pathway independently triggers apoptosis and a Rac1/Slpr/JNK/Dilp8 signaling favoring tissue homeostasis in a chronic ER stress drosophila model. Cell Death Dis. 2014, 5, e1452. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Ryoo, H.D.; Park, J.E.; Yoon, J.H.; Kang, M.J. A Drosophila reporter for the translational activation of ATF4 marks stressed cells during development. PLoS ONE 2015, 10, e0126795. [Google Scholar] [CrossRef] [PubMed]
- Sone, M.; Zeng, X.; Larese, J.; Ryoo, H.D. A modified upr stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell Stress Chaperones 2013, 18, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. CMLS 2005, 62, 670–684. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. Modomics: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006, 34, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Breton, M.; Sirand-Pugnet, P.; Tardy, F.; Thiaucourt, F.; Citti, C.; Barre, A.; Yoshizawa, S.; Fourmy, D.; de Crecy-Lagard, V.; et al. Predicting the minimal translation apparatus: Lessons from the reductive evolution of mollicutes. PLoS Genet. 2014, 10, e1004363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002, 277, 16391–16395. [Google Scholar] [CrossRef] [PubMed]
- McKenney, K.M.; Alfonzo, J.D. From prebiotics to probiotics: The evolution and functions of tRNA modifications. Life 2016, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Batlle, E.; de Pouplana, L.R. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Ignatova, Z. Emerging roles of trna in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- St Pierre, S.E.; Ponting, L.; Stefancsik, R.; McQuilton, P.; FlyBase, C. Flybase 102—Advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014, 42, D780–D788. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Cully, M.; Andersen, D.; Leevers, S.J. Insulin delays the progression of Drosophila cells through G2/M by activating the dTOR/dRaptor complex. EMBO J. 2007, 26, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Duechler, M.; Leszczynska, G.; Sochacka, E.; Nawrot, B. Nucleoside modifications in the regulation of gene expression: Focus on trna. Cell. Mol. Life Sci. CMLS 2016, 73, 3075–3095. [Google Scholar] [CrossRef] [PubMed]
- Endres, L.; Dedon, P.C.; Begley, T.J. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015, 12, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.; Pang, Y.L.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Grunewald, P.; Thuring, K.L.; Eichler, C.; Helm, M.; Schaffrath, R. Loss of anticodon wobble uridine modifications affects tRNAlys function and protein levels in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0119261. [Google Scholar] [CrossRef] [PubMed]
- Zinshteyn, B.; Gilbert, W.V. Loss of a conserved trna anticodon modification perturbs cellular signaling. PLoS Genet. 2013, 9, e1003675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullor, J.L.; Guerrero, I. A gain-of-function mutant of patched dissects different responses to the hedgehog gradient. Dev. Biol. 2000, 228, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.; Glavic, A.; Casado, M.; de Celis, J.F. A gain-of-function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 2009, 183, 1005–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | Expression Pattern | Reference |
|---|---|---|
| tubulin (tub > Gal4) | Ubiquitous | BDSC (5138) |
| engrailed (en > Gal4) | Posterior compartment | BDSC (1973) |
| hedgehog (hh > Gal4) | Posterior compartment | Mullor et al. [55] |
| collagen type IV (cg > Gal) | Fat body (larval tissue) | BDSC (7011) |
| nubbin (nub > Gal4) | Wing pouch (imaginal disc) | BDSC (42699) |
| Name | Utility | Reference |
|---|---|---|
| UAS-Tcs2 IR | tcs2 knockdown | VDRC (dna13368) |
| UAS-Tcs3 IR | tcs3 knockdown | VDRC (106250) |
| UAS-Tcs5 IR | tcs5 knockdown | VDRC (dna7059) |
| UAS-Tcs6 IR | tcs6 knockdown | VDRC (4371) |
| UAS-GFP | GFP expression | BDSC (5137) |
| UAS-Tcs3 | Tcs3 expression | Rojas-Benitez et al. [12] |
| UAS-p35 | p35 expression | BDSC (5072) |
| UAS-BSKDN | Dominant negative BSK expression | BDSC (6409) |
| GFP-IR | GFP knockdown | BDSC (9331) |
| tub > Atf4 5’UTR::dsRed | UPR activation reporter | Kang et al. [36] |
| UAS-Xbp1::GFP | UPR activation reporter | Sone et al. [37] |
| UAS-Hsc70 | Hsc70 expression | BDSC (5843) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas‐Benítez, D.; Eggers, C.; Glavic, A. Modulation of the Proteostasis Machinery to Overcome Stress Caused by Diminished Levels of t6A‐Modified tRNAs in Drosophila. Biomolecules 2017, 7, 25. https://doi.org/10.3390/biom7010025
Rojas‐Benítez D, Eggers C, Glavic A. Modulation of the Proteostasis Machinery to Overcome Stress Caused by Diminished Levels of t6A‐Modified tRNAs in Drosophila. Biomolecules. 2017; 7(1):25. https://doi.org/10.3390/biom7010025
Chicago/Turabian StyleRojas‐Benítez, Diego, Cristián Eggers, and Alvaro Glavic. 2017. "Modulation of the Proteostasis Machinery to Overcome Stress Caused by Diminished Levels of t6A‐Modified tRNAs in Drosophila" Biomolecules 7, no. 1: 25. https://doi.org/10.3390/biom7010025





