QueF-Like, a Non-Homologous Archaeosine Synthase from the Crenarchaeota
Abstract
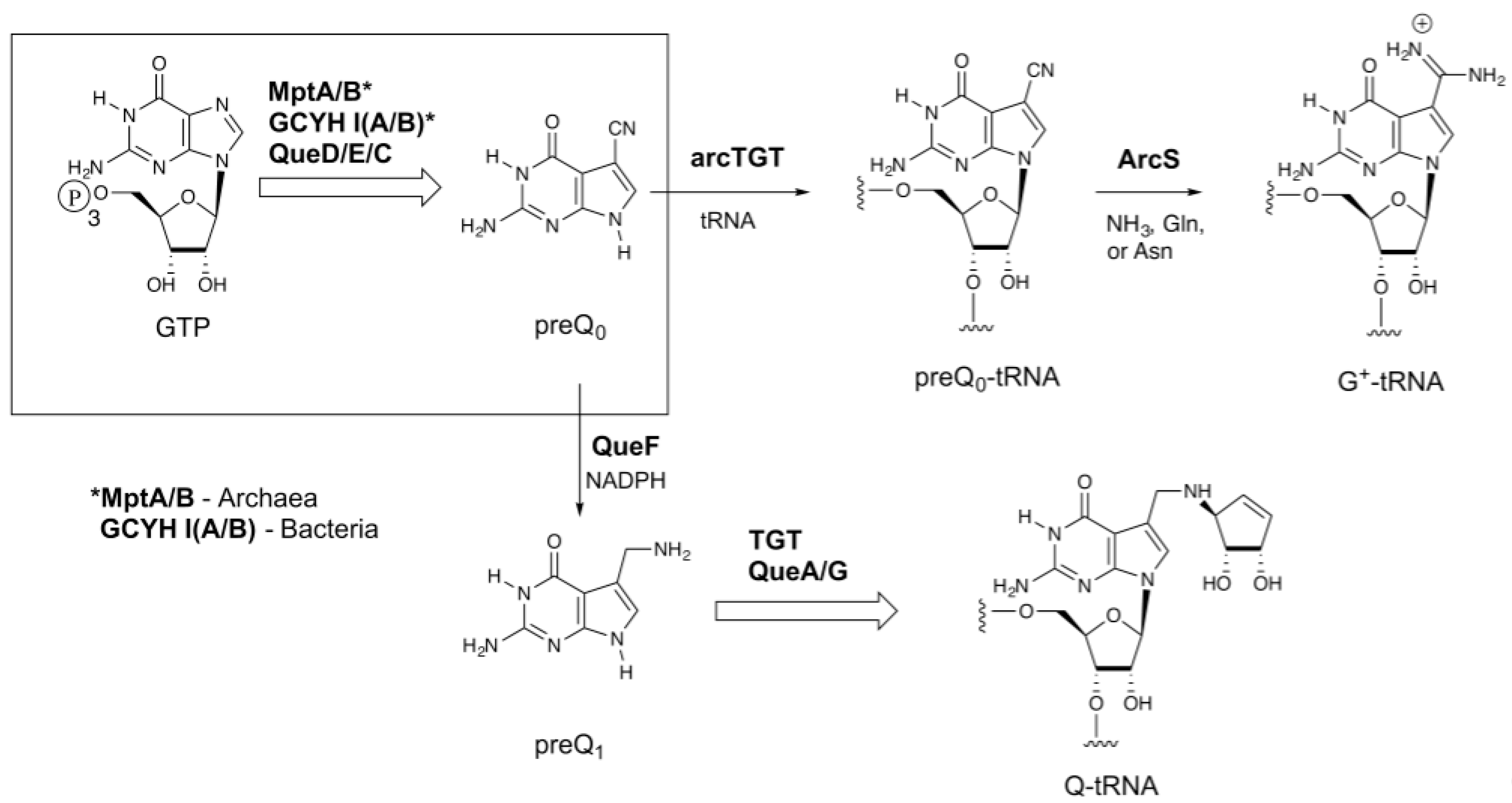
:1. Introduction
2. Results
2.1. Over-Expression of queF-L and Purification of Recombinant Pyrobaculum calidifontis QueF-L
2.2. Identification of the Substrates for P. calidifontis QueF-L
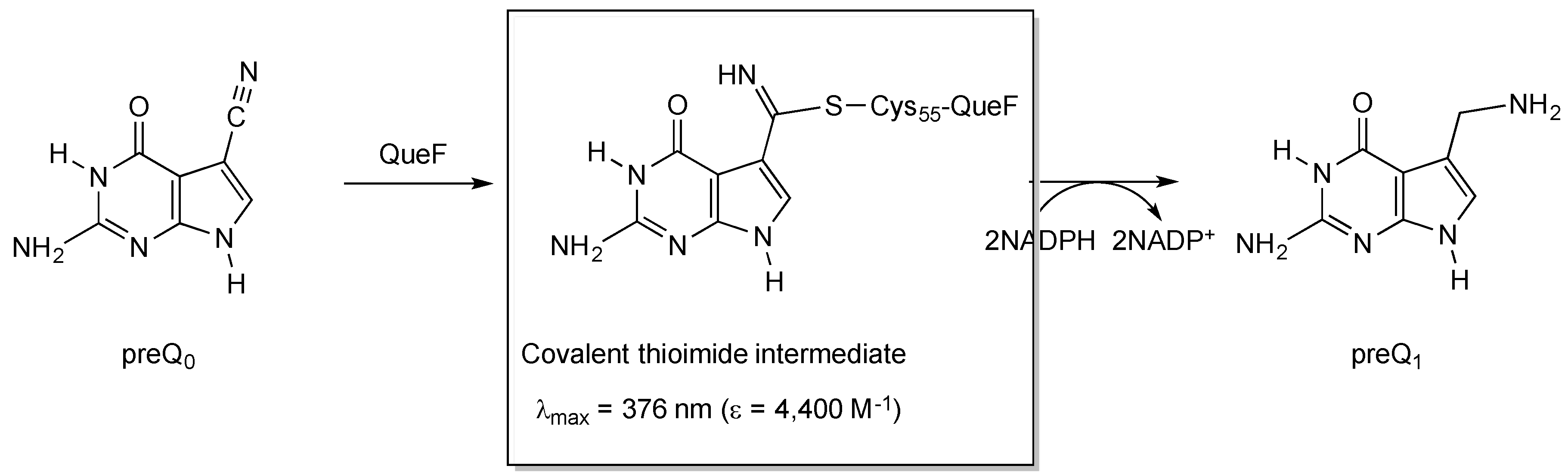
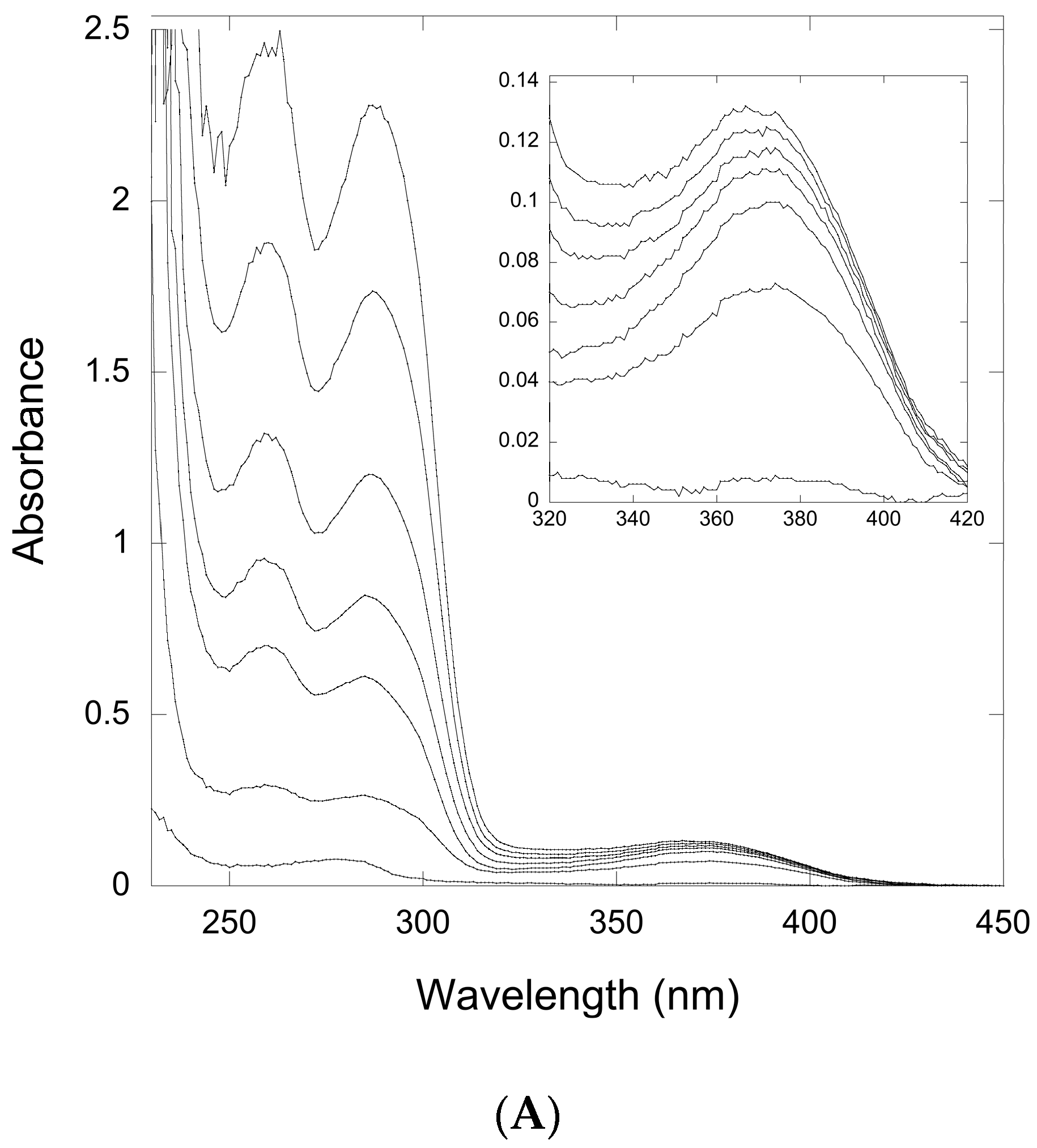
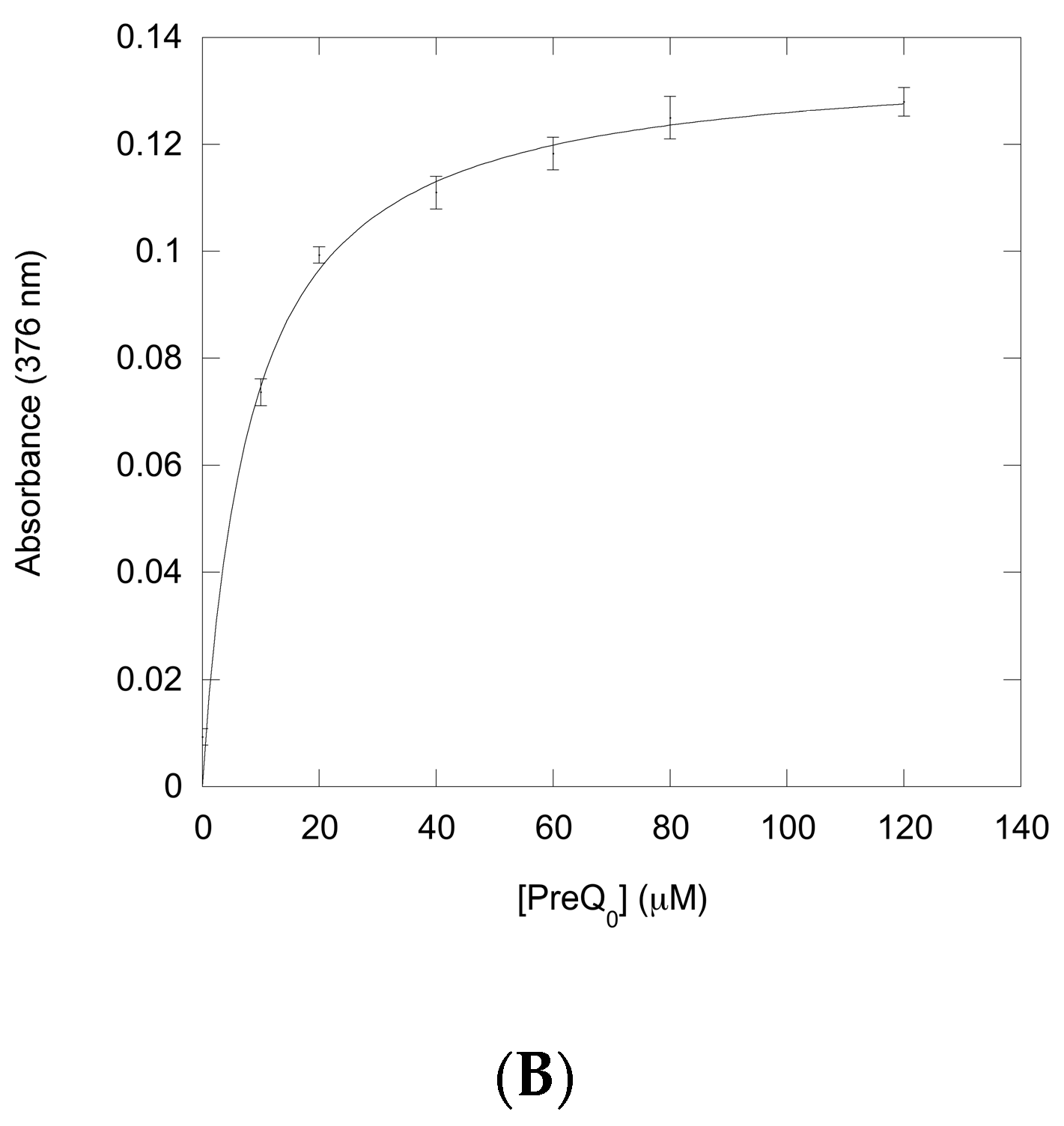
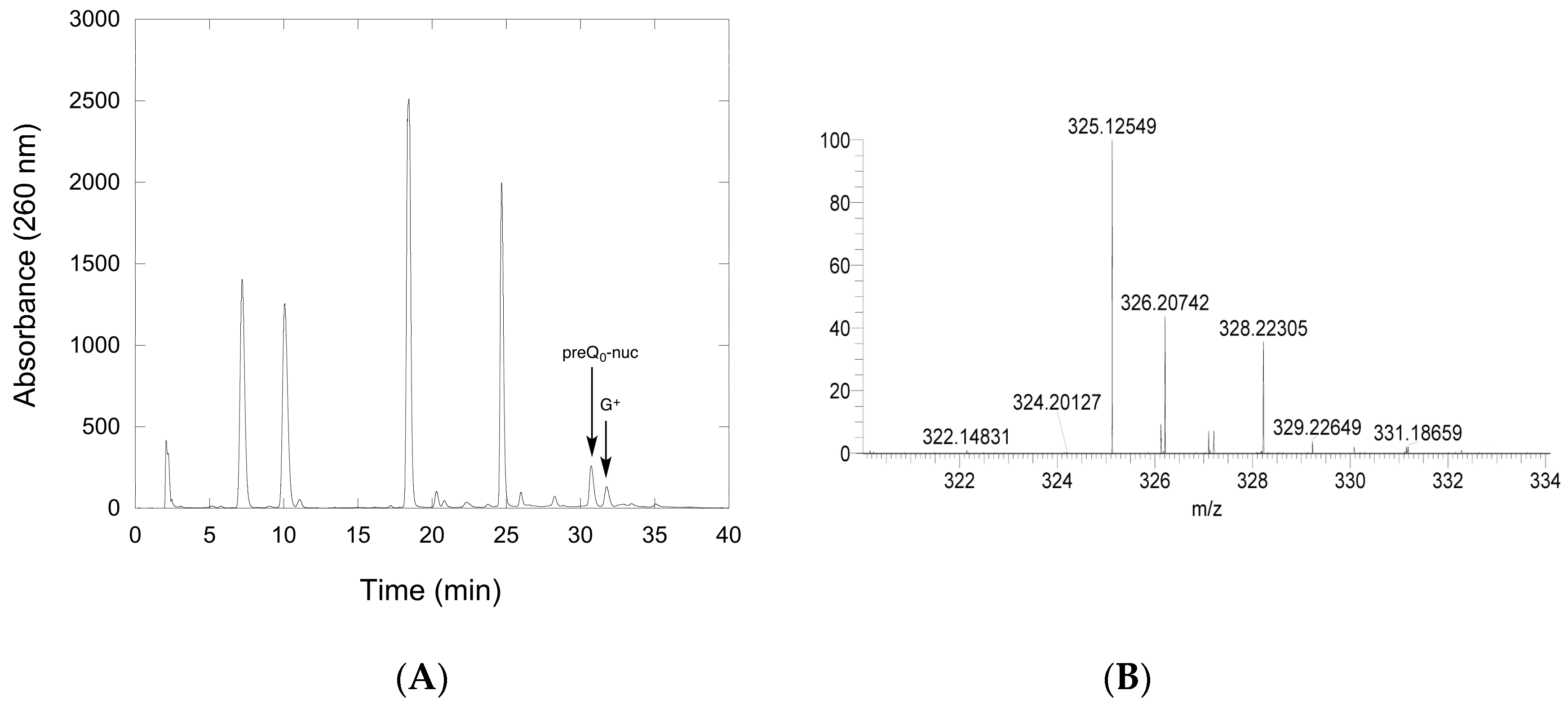
2.2.1. Thioimide Formation with preQ0 and preQ0-tRNA
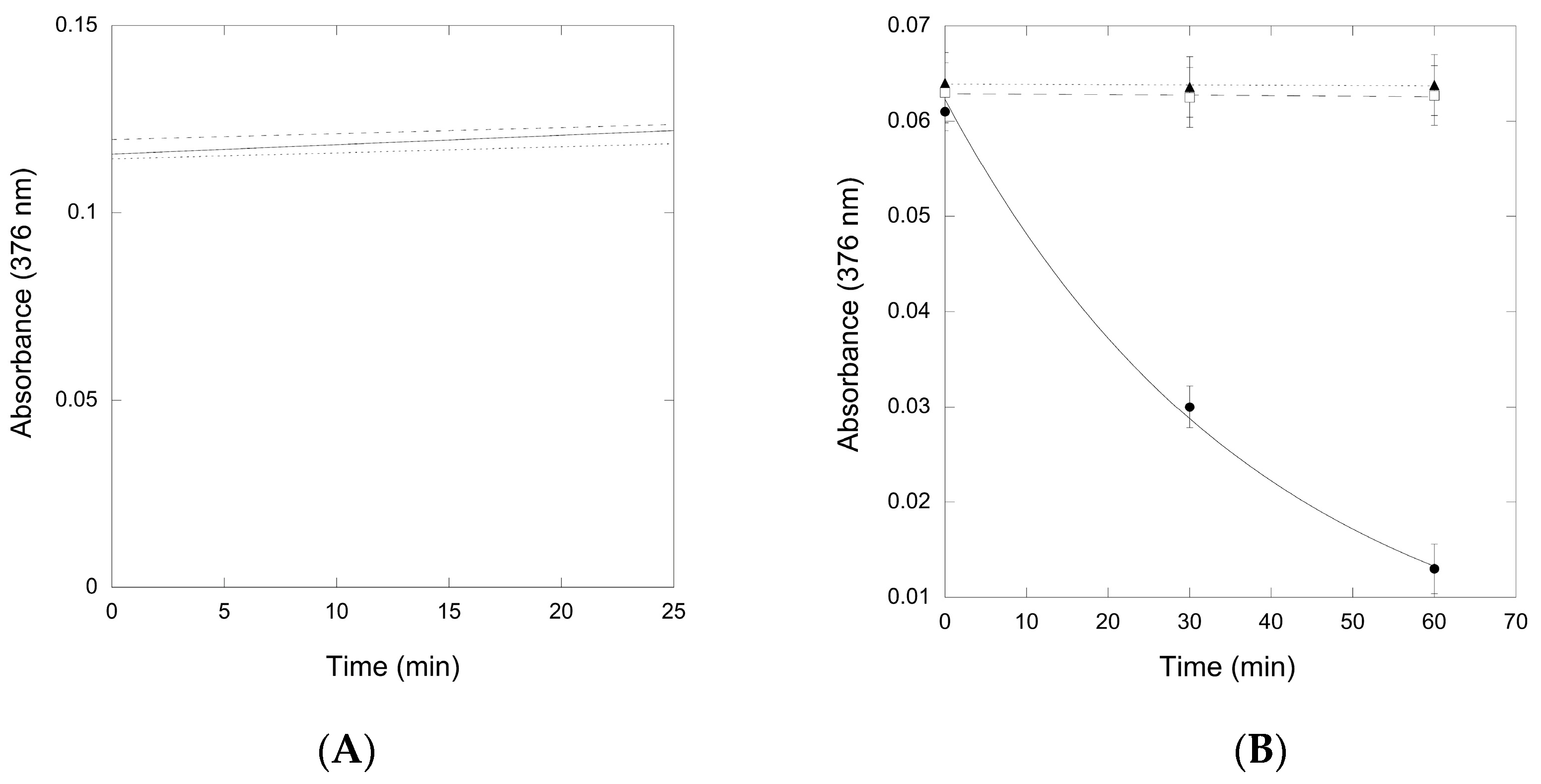
2.2.2. Reactivity of preQ0 and preQ0-tRNA Thioimides with Various Nitrogen Sources
2.2.3. Conversion of preQ0-Modified tRNA to G+-Modified tRNA by QueF-L
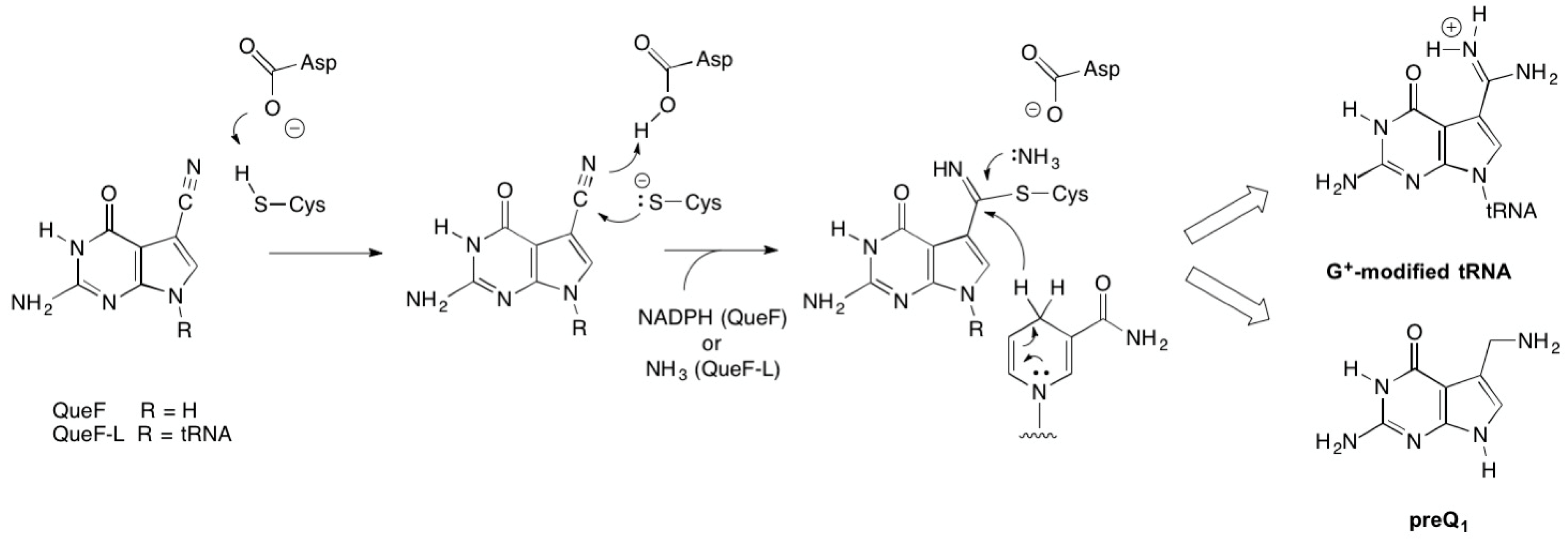
3. Discussion
4. Materials and Methods
4.1. General
4.2. Enzymes
4.3. Instrumentation
4.4. PreQ0 Synthesis
4.5. Cloning of P. calidifontis queF-L
- Sense primer: 5′-GGTATTGAGGGTCGCATGCTGAAAGTCTCAAAAAGCC-3′
- Antisense primer: 5′-AGAGGAGAGTTAGAGCCTTAGATGTAGACCGGCGGC-3′
4.6. Over-Production and Purification of Recombinant P. calidifontis QueF-L
4.7. In Vitro Transcription of tRNA
- 5′-GCAGTAATACGACTCACTATAGGTCCCGTGGGGTAGTGGTAATCCTGCTGGGCTTTG-3′
- 5′-TGGTAGTCCCGAGCGGAGTCGAACCGCTGTCGCCGGGTCCAAAGCCCAGC-3′
4.8. Preparation of preQ0 modified tRNAGln
4.9. Guanine Incorporation Controls
4.10. Substrate Titration Studies
4.11. Amidinotransferase Assays
4.12. LCMS Analysis of QueF-L Assays with preQ0-tRNA
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H. Nucleic acids are not boring long polymers of only four types of nucleosides: A guided tour. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution; Grosjean, H., Ed.; Landes Bioscience: Austin, TX, USA, 2009; pp. 1–18. [Google Scholar]
- Iwata-Reuyl, D.; de Crécy Lagard, V. Enzymatic formation of the 7-deazaguanosine hypermodified nucleosides of tRNA. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution; Grosjean, H., Ed.; Landes Bioscience: New York, NY, USA, 2009; pp. 379–394. [Google Scholar]
- Gregson, J.M.; Crain, P.F.; Edmonds, C.G.; Gupta, R.; Hashizume, T.; Phillipson, D.W.; McCloskey, J.A. Structure of archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-b-d-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)). J. Biol. Chem. 1993, 268, 10076–10086. [Google Scholar] [PubMed]
- Kasai, H.; Ohashi, Z.; Harada, F.; Nishimura, S.; Oppenheimer, N.J.; Crain, P.F.; Liehr, J.G.; von Minden, D.L.; McCloskey, J.A. Structure of the modified nucleoside Q isolated from Escherichia coli transfer ribonucleic acid. 7-(4,5-cis-dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanosine. Biochemistry 1975, 14, 4198–4208. [Google Scholar] [CrossRef] [PubMed]
- Ohgi, T.; Kondo, T.; Goto, T. Total synthesis of optically pure nucleoside Q. Determination of absolute configuration of natural nucleoside Q. J. Am. Chem. Soc. 1979, 101, 3629–3633. [Google Scholar] [CrossRef]
- Okada, N.; Nishimura, S. Enzymatic synthesis of Q* nucleoside containing mannose in the anticodon of tRNA: Isolation of a novel mannosyltransferase from a cell-free extract of rat liver. Nucleic Acids Res. 1977, 4, 2931–2937. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Shindo-Okada, N.; Nishimura, S. Isolation of mammalian tRNAAsp and tRNATyr by lectin-sepharose affinity column chromatography. Nucleic Acids Res. 1977, 4, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.C.; Ambrogelly, A.; Crain, P.F.; McCloskey, J.A.; Soll, D. A truncated aminoacyl-tRNA synthetase modifies RNA. Proc. Natl. Acad. Sci. USA 2004, 101, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Dubois, D.Y.; Blaise, M.; Becker, H.D.; Campanacci, V.; Keith, G.; Giege, R.; Cambillau, C.; Lapointe, J.; Kern, D. An aminoacyl-tRNA synthetase-like protein encoded by the Escherichia coli yadB gene glutamylates specifically tRNAAsp. Proc. Natl. Acad. Sci. USA 2004, 101, 7530–7535. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Dubois, D.Y.; Becker, H.D.; Kern, D.; Spinelli, S.; Valencia, C.; Pagot, F.; Salomoni, A.; Grisel, S.; Vincentelli, R.; et al. The Escherichia coli yadB gene product reveals a novel aminoacyl-tRNA synthetase like activity. J. Mol. Biol. 2004, 337, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Blaise, M.; Becker, H.D.; Keith, G.; Cambillau, C.; Lapointe, J.; Giege, R.; Kern, D. A minimalist glutamyl-tRNA synthetase dedicated to aminoacylation of the tRNAAsp QUC anticodon. Nucleic Acids Res. 2004, 32, 2768–2775. [Google Scholar] [CrossRef] [PubMed]
- Kersten, H. The nutrient factor queuine: Biosynthesis, occurence in transfer RNA and function. BioFactors 1988, 1, 27–29. [Google Scholar] [PubMed]
- Kersten, H.; Kersten, W. Biosynthesis and function of queuine and queuosine tRNAs. In Chromatography and Modification of Nucleosides Part B; Gehrke, C.W., Kuo, K.C.T., Eds.; Elsevier: Amsterdam, The Netherlands, 1990; pp. B69–B108. [Google Scholar]
- Marks, T.; Farkas, W.R. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem. Biophys. Res. Commun. 1997, 230, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Carlson, B.A.; Kwon, S.Y.; Chamorro, M.; Oroszlan, S.; Hatfield, D.L.; Lee, B.J. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology 1999, 255, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Durand, J.; Okada, N.; Tobe, T.; Watarai, M.; Fukuda, I.; Suzuki, T.; Nakata, N.; Komatsu, K.; Yoshikawa, M.; Sasakawa, C. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J. Bacteriol. 1994, 176, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, T.; Hirata, A.; Ohno, S.; Nomura, Y.; Nagano, T.; Nameki, N.; Yokogawa, T.; Hori, H. Multisite-specific archaeosine tRNA-guanine transglycosylase (ArcTGT) from Thermoplasma acidophilum, a thermo-acidophilic archaeon. Nucleic Acids Res. 2016, 44, 1894–1908. [Google Scholar] [CrossRef] [PubMed]
- Sprinzl, M.; Hartmann, T.; Weber, J.; Blank, J.; Zeidler, R. Compilation of tRNA sequences and sequences of tRNA genes. Nuc. Acids Res. 1989, 26, 148–153. [Google Scholar] [CrossRef]
- Oliva, R.; Tramontano, A.; Cavallo, L. Mg2+ binding and archaeosine modification stabilize the G15–C48 Levitt base pair in tRNAs. RNA 2007, 13, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Mashhadi, Z.; Xu, H.; White, R.H. An Fe2+-dependent cyclic phosphodiesterase catalyzes the hydrolysis of 7,8-dihydro-d-neopterin 2’,3’-cyclic phosphate in methanopterin biosynthesis. Biochemistry 2009, 48, 9384–9392. [Google Scholar] [CrossRef] [PubMed]
- Grochowski, L.L.; Xu, H.; Leung, K.; White, R.H. Characterization of an Fe2+-dependent archaeal-specific GTP cyclohydrolase, MptA, from Methanocaldococcus jannaschii. Biochemistry 2007, 46, 6658–6667. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.; El Yacoubi, B.; Lyons, B.; Alvarez, S.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: A new role for GTP cyclohydrolase I. J. Bacteriol. 2008, 190, 7876–7884. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bonnett, S.; Anderson, J.N.; Swairjo, M.A.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 2006, 281, 37586–37593. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.; Chikwana, V.M.; Maxwell, A.; El-Yacoubi, B.; Swairjo, M.A.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Discovery and characterization of an amidinotransferase involved in the modification of archaeal tRNA. J. Biol. Chem. 2010, 285, 12706–12713. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.; Swairjo, M.A.; Gaston, K.W.; Bailly, M.; Limbach, P.A.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Diversity of archaeosine synthesis in Crenarchaeota. ACS Chem. Biol. 2012, 7, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Van Lanen, S.G.; Reader, J.S.; Swairjo, M.A.; de Crecy-Lagard, V.; Lee, B.; Iwata-Reuyl, D. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. Proc. Natl. Acad. Sci. USA 2005, 102, 4264–4269. [Google Scholar] [CrossRef] [PubMed]
- Chikwana, V.M.; Stec, B.; Lee, B.W.; de Crecy-Lagard, V.; Iwata-Reuyl, D.; Swairjo, M.A. Structural basis of biological nitrile reduction. J. Biol. Chem. 2012, 287, 30560–30570. [Google Scholar] [CrossRef] [PubMed]
- Colloc’h, N.; Poupon, A.; Mornon, J.P. Sequence and structural features of the T-fold, an original tunnelling building unit. Proteins 2000, 39, 142–154. [Google Scholar] [CrossRef]
- Mei, X.; Alvarez, J.; Bon Ramos, A.; Samanta, U.; Iwata-Reuyl, D.; Swairjo, M.A. Crystal structure of the archaeosine synthase QueF-Like—Insights into amidino transfer and tRNA recognition by the tunnel fold. Proteins 2017, 85, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Matsuo, M.; Tanaka, S.; Akimoto, H.; Asahi, S.; Nishimura, S.; Katz, J.R.; Hashizume, T.; Crain, P.F.; McCloskey, J.A.; et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement of the tRNA polynucleotide chain. J. Biol. Chem. 1997, 272, 20146–20151. [Google Scholar] [CrossRef] [PubMed]
- Hoops, G.C.; Townsend, L.B.; Garcia, G.A. tRNA-guanine transglycosylase from Escherichia coli: Structure-activity studies investigating the role of the aminomethyl substituent of the heterocyclic substrate PreQ1. Biochemistry 1995, 34, 15381–15387. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, R.; Nureki, O.; Fukai, S.; Kijimoto, T.; Nameki, N.; Watanabe, M.; Kondo, H.; Sekine, M.; Okada, N.; Nishimura, S.; et al. Crystal structure of archaeosine tRNA-guanine transglycosylase. J. Mol. Biol. 2002, 318, 665–677. [Google Scholar] [CrossRef]
- Romier, C.; Meyer, J.E.; Suck, D. Slight sequence variations of a common fold explain the substrate specificities of tRNA-guanine transglycosylases from the three kingdoms. FEBS Lett. 1997, 416, 93–98. [Google Scholar] [CrossRef]
- Lee, B.W.; van Lanen, S.G.; Iwata-Reuyl, D. Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis. Biochemistry 2007, 46, 12844–12854. [Google Scholar] [CrossRef] [PubMed]
- Massiere, F.; Badet-Denisot, M.A. The mechanism of glutamine-dependent amidotransferases. Cell. Mol. Life Sci. 1998, 54, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Zalkin, H. The amidotransferases. Adv. Enzymol. Relat. Areas Mol. Biol. 1993, 66, 203–309. [Google Scholar] [PubMed]
- Zallot, R.; Ross, R.; Chen, W.H.; Bruner, S.D.; Limbach, P.A.; De Crecy-Lagard, V. Identification of a novel epoxyqueuosine reductase family by comparative genomics. ACS Chem. Biol. 2017, 12, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Miles, Z.D.; McCarty, R.M.; Molnar, G.; Bandarian, V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc. Natl. Acad. Sci. USA 2011, 108, 7368–7372. [Google Scholar] [CrossRef] [PubMed]
- Lyakhov, D.L.; He, B.; Zhang, X.; Studier, F.W.; Dunn, J.J.; McAllister, W.T. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J. Mol. Biol. 1997, 269, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Fox, D.T.; Lacy, J.A.; Van Lanen, S.G.; Iwata-Reuyl, D. Hypermodification of tRNA in Thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J. Biol. Chem. 2000, 275, 28731–28738. [Google Scholar] [CrossRef] [PubMed]
- Sherlin, L.D.; Bullock, T.L.; Nissan, T.A.; Perona, J.J.; Lariviere, F.J.; Uhlenbeck, O.C.; Scaringe, S.A. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA 2001, 7, 1671–1678. [Google Scholar] [PubMed]
- Bhaskaran, H.; Rodriguez-Hernandez, A.; Perona, J.J. Kinetics of tRNA folding monitored by aminoacylation. RNA 2012, 18, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Crain, P.F. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Meth. Enzymol. 1990, 193, 782–790. [Google Scholar] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bon Ramos, A.; Bao, L.; Turner, B.; De Crécy-Lagard, V.; Iwata-Reuyl, D. QueF-Like, a Non-Homologous Archaeosine Synthase from the Crenarchaeota. Biomolecules 2017, 7, 36. https://doi.org/10.3390/biom7020036
Bon Ramos A, Bao L, Turner B, De Crécy-Lagard V, Iwata-Reuyl D. QueF-Like, a Non-Homologous Archaeosine Synthase from the Crenarchaeota. Biomolecules. 2017; 7(2):36. https://doi.org/10.3390/biom7020036
Chicago/Turabian StyleBon Ramos, Adriana, Lide Bao, Ben Turner, Valérie De Crécy-Lagard, and Dirk Iwata-Reuyl. 2017. "QueF-Like, a Non-Homologous Archaeosine Synthase from the Crenarchaeota" Biomolecules 7, no. 2: 36. https://doi.org/10.3390/biom7020036





