The Extracellular Domain of Human High Affinity Copper Transporter (hNdCTR1), Synthesized by E. coli Cells, Chelates Silver and Copper Ions In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. E. coli Strain and Growth Conditions
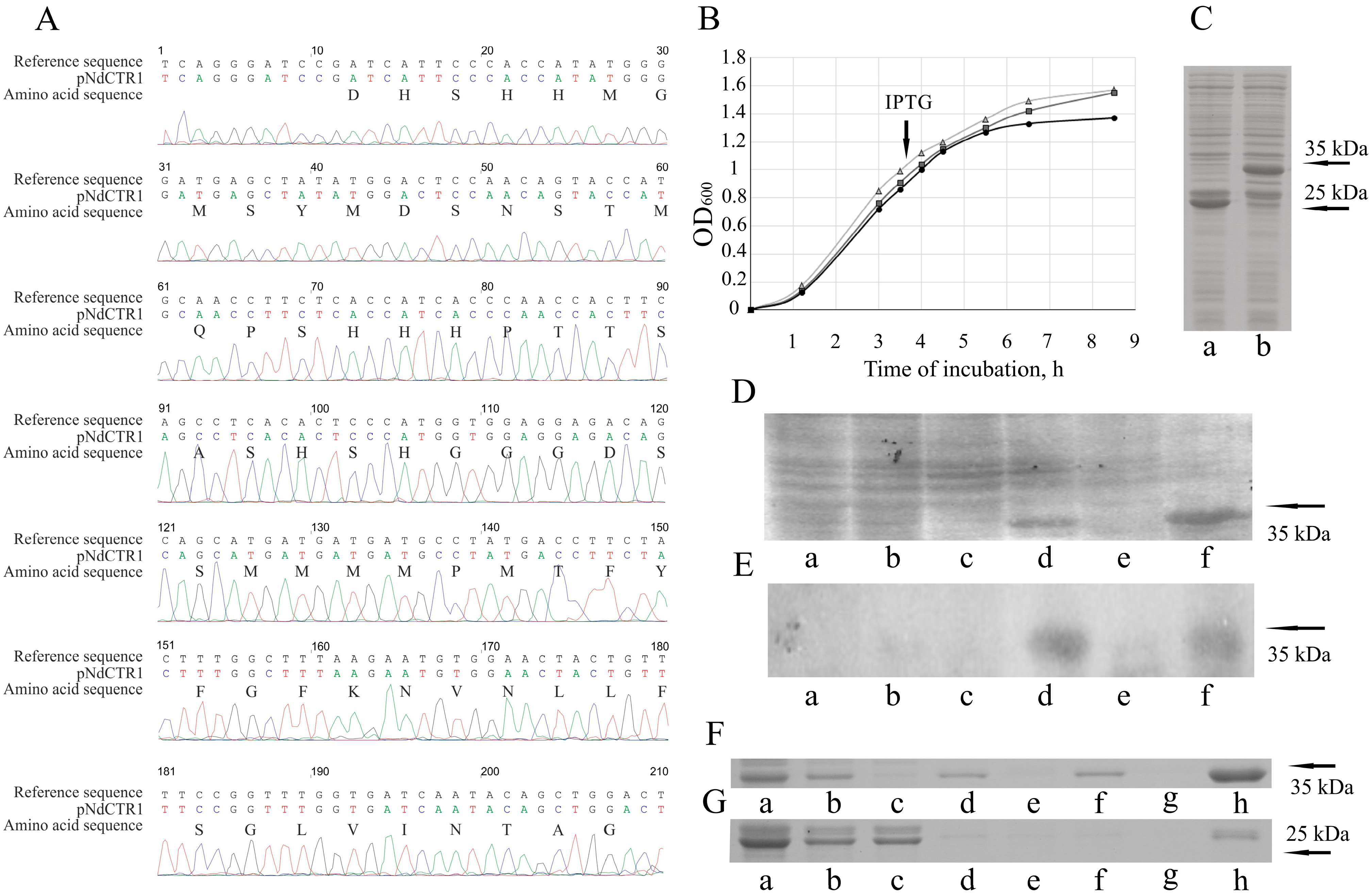
2.2. Cloning of N-Terminal Extracellular Domain of hCTR1
2.3. Polyacrylamide Gel Electrophoresis, Immunoblotting, and Immunoprecipitation
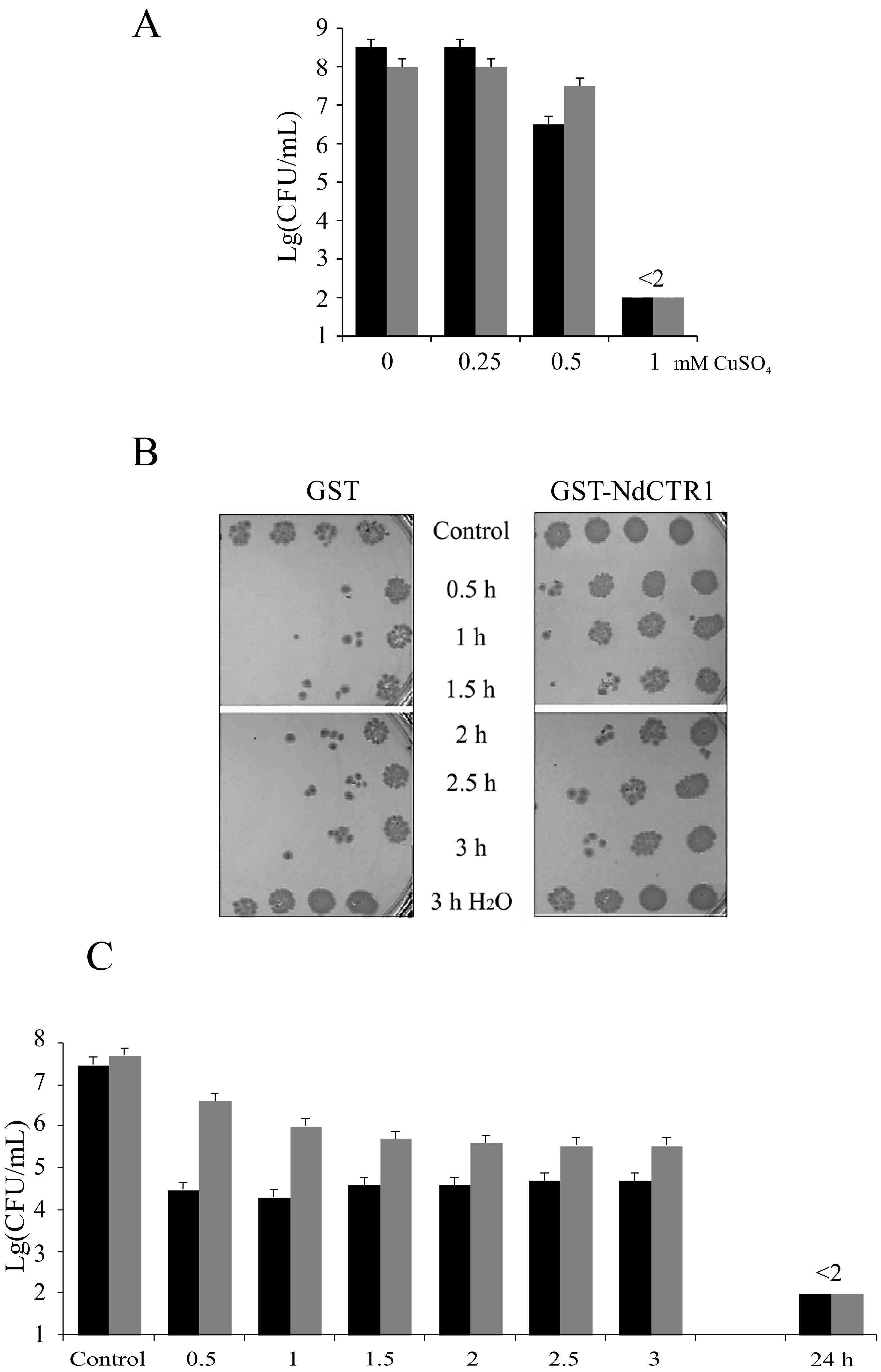
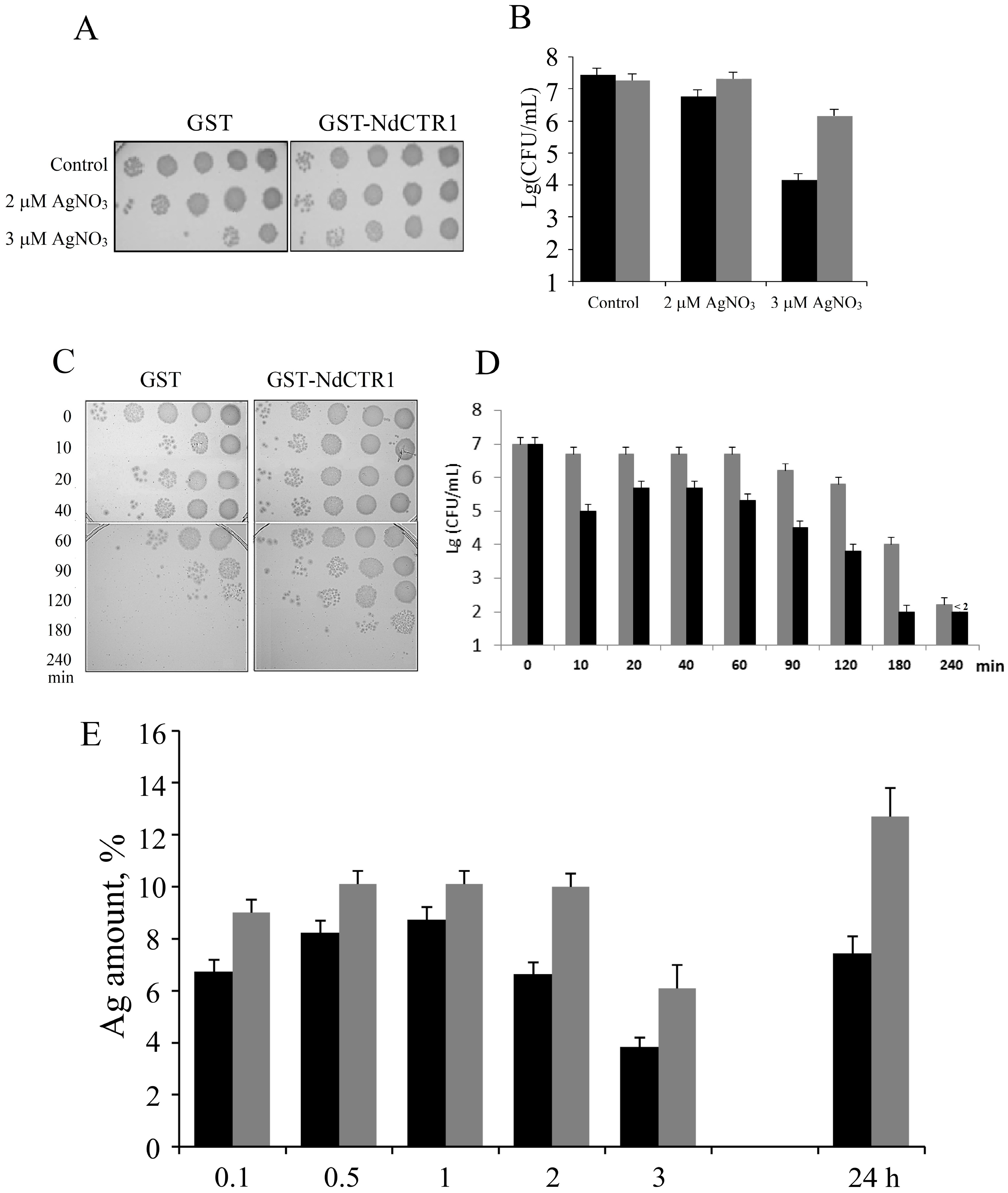
2.4. Treatment of Cells with CuSO4 or AgNO3
2.5. Disruption and Fractionation of Bacterial Cells
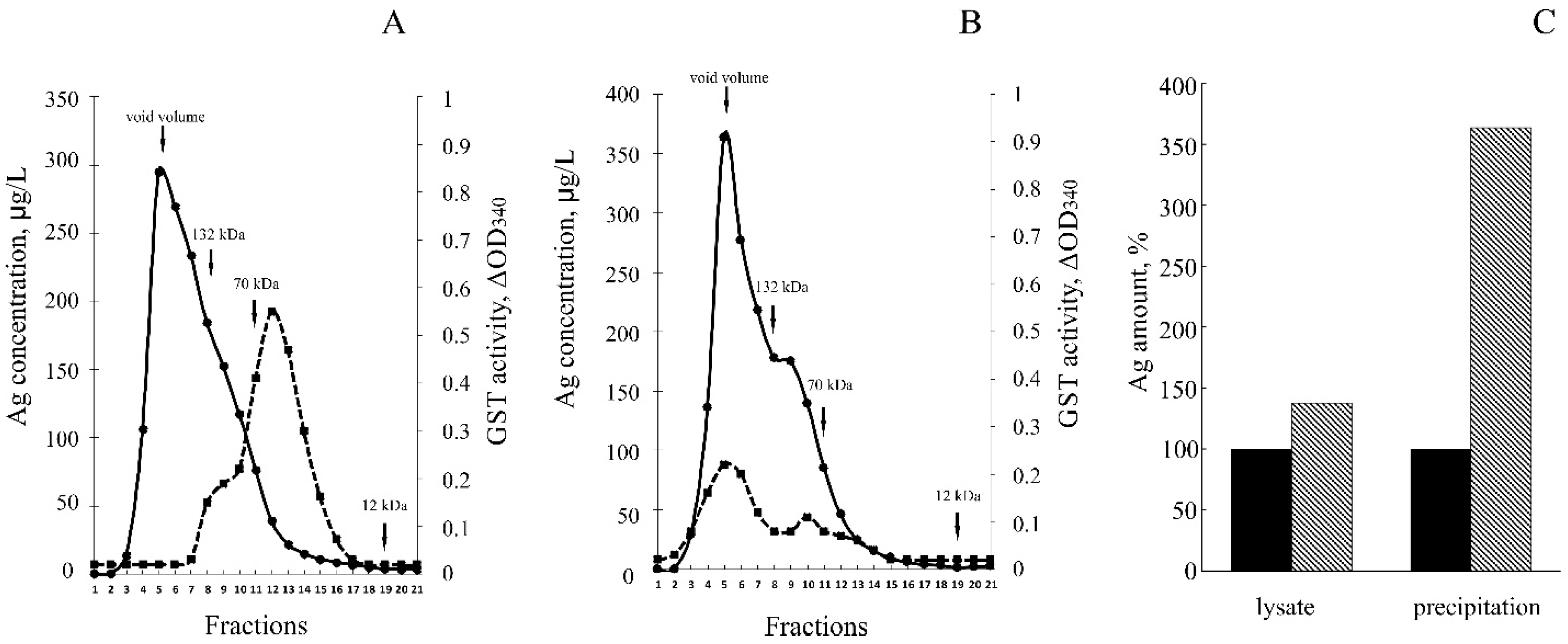
2.6. Gel-Filtration Chromatography
2.7. GST Activity Determination
2.8. Transmission Electron Microscopy Analysis of Cells
2.9. Measurement of Silver Concentration
3. Results
3.1. Cloning of hCTR1 Ectodomain
3.2. hNdCTR1 Polypeptide Rescues E. coli Cells from the Toxic Action of Copper and Silver Ions
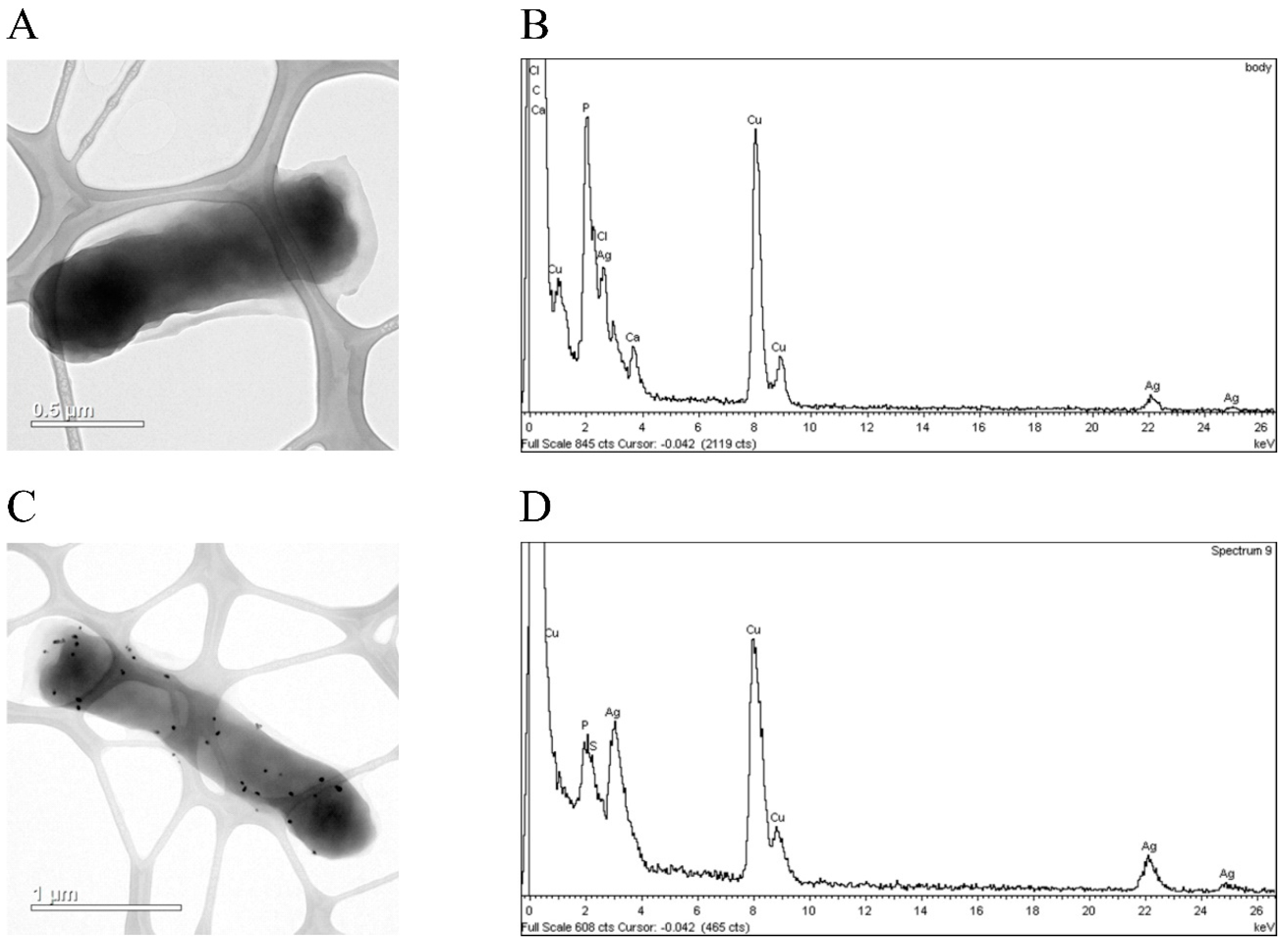
3.3. hNdCTR1 Chelates Silver Ions
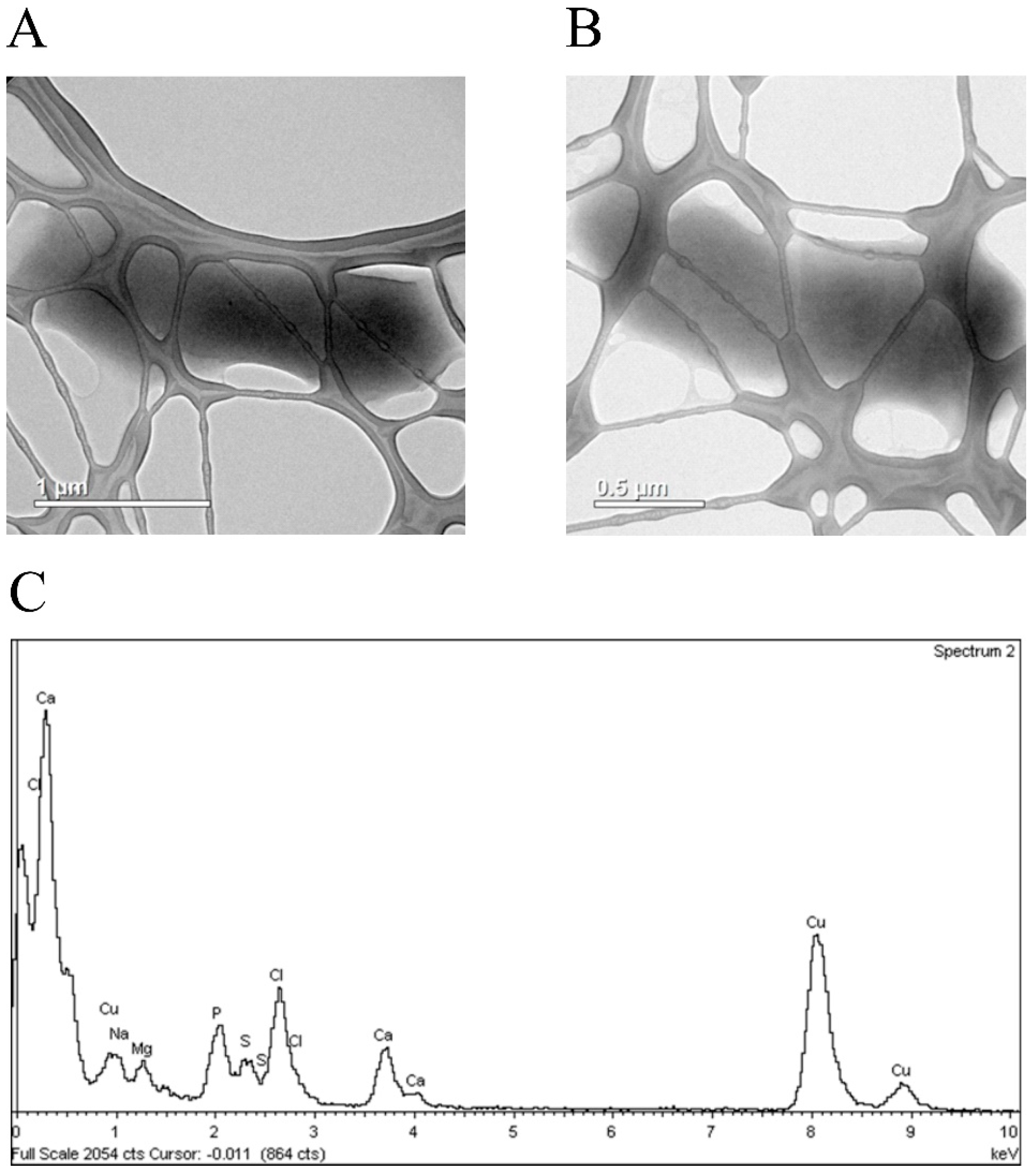
3.4. Silver Ions Induced Morphological Changes in E. coli Cells, Which Expressed Fusion Protein
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Palm-Espling, M.E.; Niemiec, M.S.; Wittung-Stafshede, P. Role of metal in folding and stability of copper proteins in vitro. Biochim. Biophys. Acta 2012, 1823, 1594–1603. [Google Scholar] [CrossRef] [PubMed]
- Tapiero, H.; Townsend, D.M.; Tew, K.D. Trace elements in human physiology and pathology. Copper Biomed. Pharmacother. 2003, 57, 386–398. [Google Scholar] [CrossRef]
- Krohn, K.A.; Link, J.M.; Mason, R.P. Molecular imaging of hypoxia. J. Nucl. Med. 2008, 49, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.D.; Tsai, W.B.; Lee, M.Y.; Savaraj, N.; Kuo, M.T. Specificity protein 1 sp1 oscillation is involved in copper homeostasis maintenance by regulating human high-affinity copper transporter 1 expression. Mol. Pharmacol. 2012, 81, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Arciello, M.; Longo, A.; Viscomi, C.; Capo, C.; Angeloni, A.; Rossi, L.; Balsano, C. Core domain mutant Y220C of p53 protein has a key role in copper homeostasis in case of free fatty acids overload. Biometals 2015, 28, 1017–1029. [Google Scholar] [CrossRef] [PubMed]
- Mufti, A.R.; Burstein, E.; Duckett, C.S. XIAP: Cell death regulation meets copper homeostasis. Arch. Biochem. Biophys. 2007, 463, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Turski, M.L.; Thiele, D.J. New roles for copper metabolism in cell proliferation; signaling; and disease. J. Biol. Chem. 2009, 284, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Ioannoni, R.; Beaudoin, J.; Lopez-Maury, L.; Codlin, S.; Bahler, J.; Labbe, S. Cuf2 is a novel meiosis-specific regulatory factor of meiosis maturation. PLoS ONE 2012, 7, e36338. [Google Scholar] [CrossRef] [PubMed]
- Turski, M.L.; Brady, D.C.; Kim, H.J.; Kim, B.E.; Nose, Y.; Counter, C.M.; Winge, D.R.; Thiele, D.J. A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol. Cell Biol. 2012, 32, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Opazo, K.M.; Greenough, M.A.; Bush, A.I. Copper: From neurotransmission to neuroproteostasis. Front. Aging Neurosci. 2014, 6, 143. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Crowe, M.S.; Turski, M.L.; Hobbs, G.A.; Yao, X.; Chaikuad, A.; Knapp, S.; Xiao, K.; Campbell, S.L.; Thiele, D.J.; et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 2014, 509, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Easter, R.N.; Qilin, C.; Lai, B.; Ritman, E.L.; Caruso, J.A.; Zhenyu, Q. Vascular metallomics: Copper in the vasculature. Vasc. Med. 2010, 15, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, H.; Kozlkowska, P.; Watly, J.; Krzywoszynska, K.; Potocki, S. General aspects of metal toxicity. Curr. Med. Chem. 2014, 21, 3721–3740. [Google Scholar] [CrossRef] [PubMed]
- Puig, S.; Lee, J.; Lau, M.; Thiele, D.J. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 2002, 277, 26021–26030. [Google Scholar] [CrossRef] [PubMed]
- Wooton-Kee, C.R.; Jain, A.K.; Wagner, M.; Grusak, M.A.; Finegold, M.J.; Lutsenko, S.; Moore, D.D. Elevated copper impairs hepatic nuclear receptor function in Wilson’s disease. J. Clin. Investig. 2015, 125, 3449–3460. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Takano, H.; Shimada, A.; Satoh, M. Metallothionein as an anti-inflammatory mediator. Mediat. Inflamm. 2009, 2009, 101659. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Lutsenko, S. Human copper transporters: Mechanism; role in human diseases and therapeutic potential. Future Med. Chem. 2009, 1, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Fujisawa, C.; Bhadhprasit, W. Inherited copper transport disorders: Biochemical mechanisms; diagnosis; and treatment. Curr. Drug Metab. 2012, 13, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Mitra, J.; Guerrero, E.N.; Hegde, P.M.; Wang, H.; Boldogh, I.; Rao, K.S.; Mitra, S.; Hegde, M.L. New perspectives on oxidized genome damage and repair inhibition by pro-oxidant metals in neurological diseases. Biomolecules 2014, 4, 678–703. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, P.; Gottlieb, S.H.; Culotta, V.L.; Navas-Acien, A. EDTA chelation therapy to reduce cardiovascular events in persons with diabetes. Curr. Cardiol. Rep. 2015, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics 2015, 7, 1459–1476. [Google Scholar] [CrossRef] [PubMed]
- Öhrvik, H.; Thiele, D.J. How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann. N. Y. Acad. Sci. 2014, 1314, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Parr, R.G.; Pearson, R.G. Absolute hardness: Companion parameter to absolute electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
- Skvortsov, A.N.; Zatulovskiĭ, E.A.; Puchkova, L.V. Structure-functional organization of eukaryotic high-affinity copper importer CTR1 determines its ability to transport copper; silver and cisplatin. Mol. Biol. Mosk. 2012, 46, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, H.; Wang, X.; Liu, Q.; Ni, J.; Sun, H. Kinetics and thermodynamics of metal binding to the N-terminus of a human copper transporter; hCTR1. Chem. Commun. Camb. 2013, 49, 9134–9136. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, X.; Li, H.; Sun, H. Comparison between copper and cisplatin transport mediated by human copper transporter 1 hCTR1. Metallomics 2012, 4, 679–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilyechova, E.Y.; Saveliev, A.N.; Skvortsov, A.N.; Babich, P.S.; Zatulovskaia, A.Y.; Pliss, M.G.; Korzhevskii, D.E.; Tsymbalenko, N.V.; Puchkova, L.V. The effects of silver ions on copper metabolism in rats. Metallomics 2014, 6, 1970–1987. [Google Scholar] [CrossRef] [PubMed]
- Babich, P.S.; Skvortsov, A.N.; Rusconi, P.; Tsymbalenko, N.V.; Mutanen, M.; Puchkova, L.V.; Broggini, M. Non-hepatic tumors change the activity of genes encoding copper trafficking proteins in the liver. Cancer Biol. Ther. 2013, 14, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Kovács, D.; Szőke, K.; Igaz, N.; Spengler, G.; Molnár, J.; Tóth, T.; Madarász, D.; Rázga, Z.; Kónya, Z.; Boros, I.M.; et al. Silver nanoparticles modulate ABC transporter activity and enhance chemotherapy in multidrug resistant cancer. Nanomedicine 2016, 12, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Sinani, D.; Adle, D.J.; Kim, H.; Lee, J. Distinct mechanisms for CTR1-mediated copper and cisplatin transport. J. Biol. Chem. 2007, 282, 26775–26785. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Wu, Q.; Wu, X.; Arnesano, F.; Natile, G.; Sletten, E.; Liu, Y. The reaction of a platinated methionine motif of CTR1 with cysteine and histidine is dependent upon the type of precursor platinum complex. J. Inorg. Biochem. 2015, 153, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.L.; Putterman, A.B.; White, D.R.; Thiele, D.J.; Franz, K.J. Model peptides provide new insights into the role of histidine residues as potential ligands in human cellular copper acquisition via Ctr1. J. Am. Chem. Soc. 2011, 133, 4427–4437. [Google Scholar] [CrossRef] [PubMed]
- Zatulovskiy, E.A.; Skvortsov, A.N.; Rusconi, P.; Ilyechova, E.Y.; Babich, P.S.; Tsymbalenko, N.V.; Broggini, M.; Puchkova, L.V. Serum depletion of holo-ceruloplasmin induced by silver ions in vivo reduces uptake of cisplatin. J. Inorg. Biochem. 2012, 116, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.; Green, L.H. Practical Handbook of Microbiology, 2nd ed.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2008. [Google Scholar]
- Sokolov, A.V.; Kostevich, V.A.; Romanico, D.N.; Zakharova, E.T.; Vasilyev, V.B. Two-stage method for purification of ceruloplasmin based on its interaction with neomycin. Biochem. Mosc. 2012, 77, 631–638. [Google Scholar] [CrossRef] [PubMed]
- De Feo, C.J.; Aller, S.G.; Siluvai, G.S.; Blackburn, N.J.; Unger, V.M. Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. USA 2009, 106, 4237–4242. [Google Scholar] [CrossRef] [PubMed]
- Argüello, J.M.; Raimunda, D.; Padilla-Benavide, T. Mechanisms of copper homeostasis in bacteria. Front. Cell. Infect. Microbiol. 2013, 3, 73. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Brose, J.; Schimo, S.; Ackland, S.M.; La Fontaine, S.; Wedd, A.G. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: Detection probes and affinity standards. J. Biol. Chem. 2011, 286, 11047–11055. [Google Scholar] [CrossRef] [PubMed]
- Jaimes-Lizcano, Y.A.; Hunn, D.D.; Papadopoulos, K.D. Filamentous Escherichia coli cells swimming in tapered microcapillaries. Res. Microbiol. 2014, 165, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.M.A.; Daniele-Silva, A.; Teixeira, D.G.; Estrela, A.B.; Melo, K.R.T.; Oliveira, V.S.; Rocha, H.A.O.; Ferreira, L.S.; Pontes, D.L.; Lima, J.P.M.S.; et al. Structure and in vitro activities of a Copper II-chelating anionic peptide from the venom of the scorpion Tityus stigmurus. Peptides 2017, 94, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Lichtmannegger, J.; Leitzinger, C.; Wimmer, R.; Schmitt, S.; Schulz, S.; Kabiri, Y.; Eberhagen, C.; Rieder, T.; Janik, D.; Neff, F.; et al. Methanobactin reverses acute liver failure in a rat model of Wilson disease. J. Clin. Investig. 2016, 126, 2721–2735. [Google Scholar] [CrossRef] [PubMed]
- De Ricco, R.; Potocki, S.; Kozlowski, H.; Valensina, D. NMR investigations of metal interactions with unstructured soluble protein domains. Coord. Chem. Rev. 2014, 269, 1–12. [Google Scholar] [CrossRef]
- Camponeschi, F.; Valensin, D.; Tessari, I.; Bubacco, L.; Dell’Acqua, S.; Casella, L.; Monzani, E.; Gaggelli, E.; Valensin, G. Copper(I)-α-synuclein Interaction: Structural description of two independent and competing metal binding sites. Inorg. Chem. 2013, 52, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Hamsell, M.A.; Xue, Y.; Robinson, C.D.; Canalizo-Hernández, M.A.; Marvin, R.G.; Kelly, R.A.; Mondragón, A.; Penner-Hahn, J.E.; O’Halloran, T.V. Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation. Science 2010, 327, 331–334. [Google Scholar] [CrossRef]
- Juling, S.; Bachler, G.; von Götz, N.; Lichtenstein, D.; Böhmert, L.; Niedzwiecka, A.; Selve, S.; Braeuning, A.; Lampen, A. In vivo distribution of nanosilver in the rat: The role of ions and de novo-formed secondary particles. Food Chem. Toxicol. 2016, 97, 327–335. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sankova, T.P.; Orlov, I.A.; Saveliev, A.N.; Kirilenko, D.A.; Babich, P.S.; Brunkov, P.N.; Puchkova, L.V. The Extracellular Domain of Human High Affinity Copper Transporter (hNdCTR1), Synthesized by E. coli Cells, Chelates Silver and Copper Ions In Vivo. Biomolecules 2017, 7, 78. https://doi.org/10.3390/biom7040078
Sankova TP, Orlov IA, Saveliev AN, Kirilenko DA, Babich PS, Brunkov PN, Puchkova LV. The Extracellular Domain of Human High Affinity Copper Transporter (hNdCTR1), Synthesized by E. coli Cells, Chelates Silver and Copper Ions In Vivo. Biomolecules. 2017; 7(4):78. https://doi.org/10.3390/biom7040078
Chicago/Turabian StyleSankova, Tatiana P., Iurii A. Orlov, Andrey N. Saveliev, Demid A. Kirilenko, Polina S. Babich, Pavel N. Brunkov, and Ludmila V. Puchkova. 2017. "The Extracellular Domain of Human High Affinity Copper Transporter (hNdCTR1), Synthesized by E. coli Cells, Chelates Silver and Copper Ions In Vivo" Biomolecules 7, no. 4: 78. https://doi.org/10.3390/biom7040078





