Loss of Suppressor of Fused in Mid-Corticogenesis Leads to the Expansion of Intermediate Progenitors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Immunohistochemistry and DiI Labeling
2.3. In Situ Hybridization
2.4. Image Acquisition and Analysis
2.5. Quantitative PCR Analysis
2.6. Western Blot Analysis
2.7. Statistics
3. Results
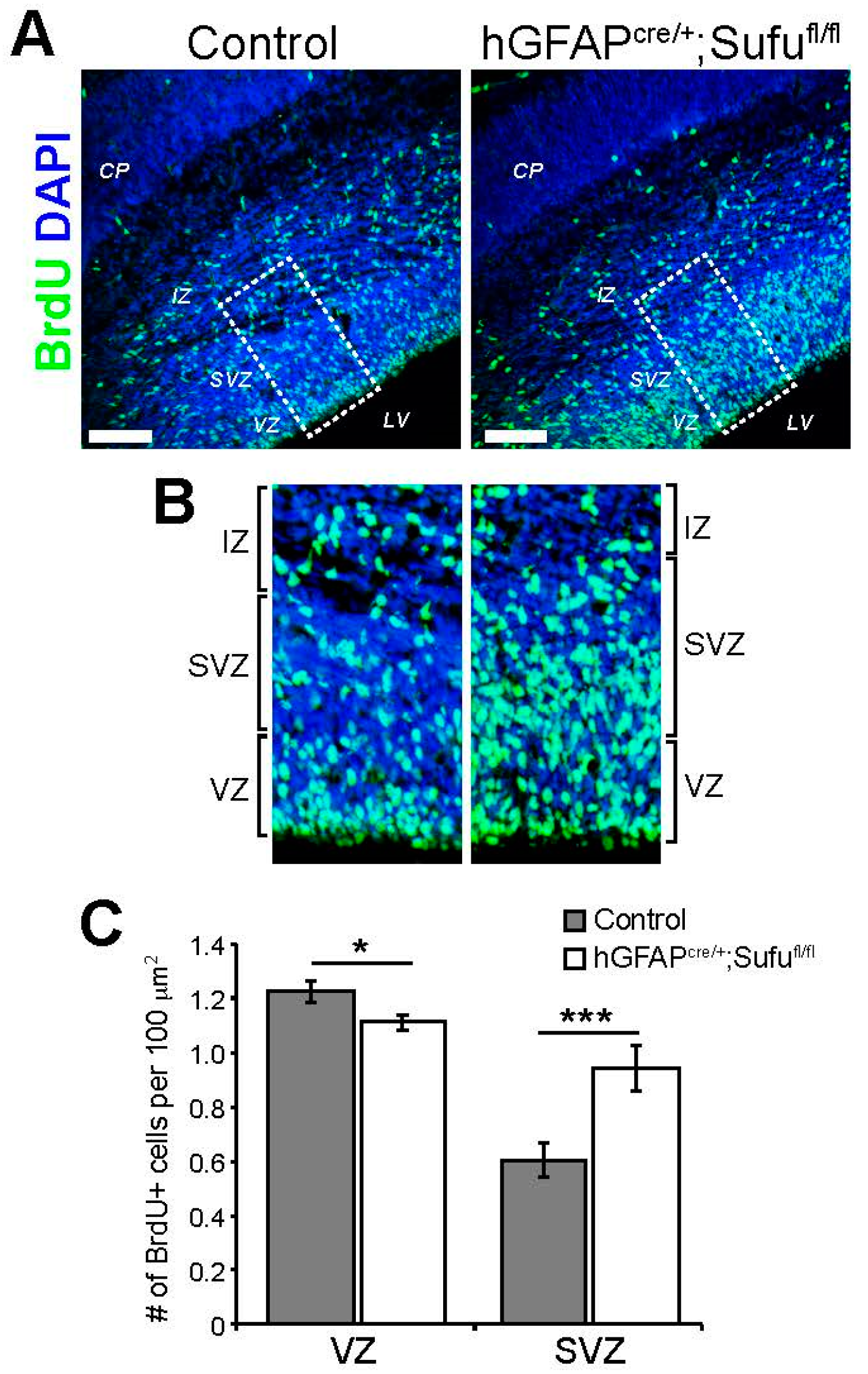
3.1. Expansion of the VZ and SVZ in the E16.5 Neocortex of hGFAPcre/+;Sufufl/fl Mice
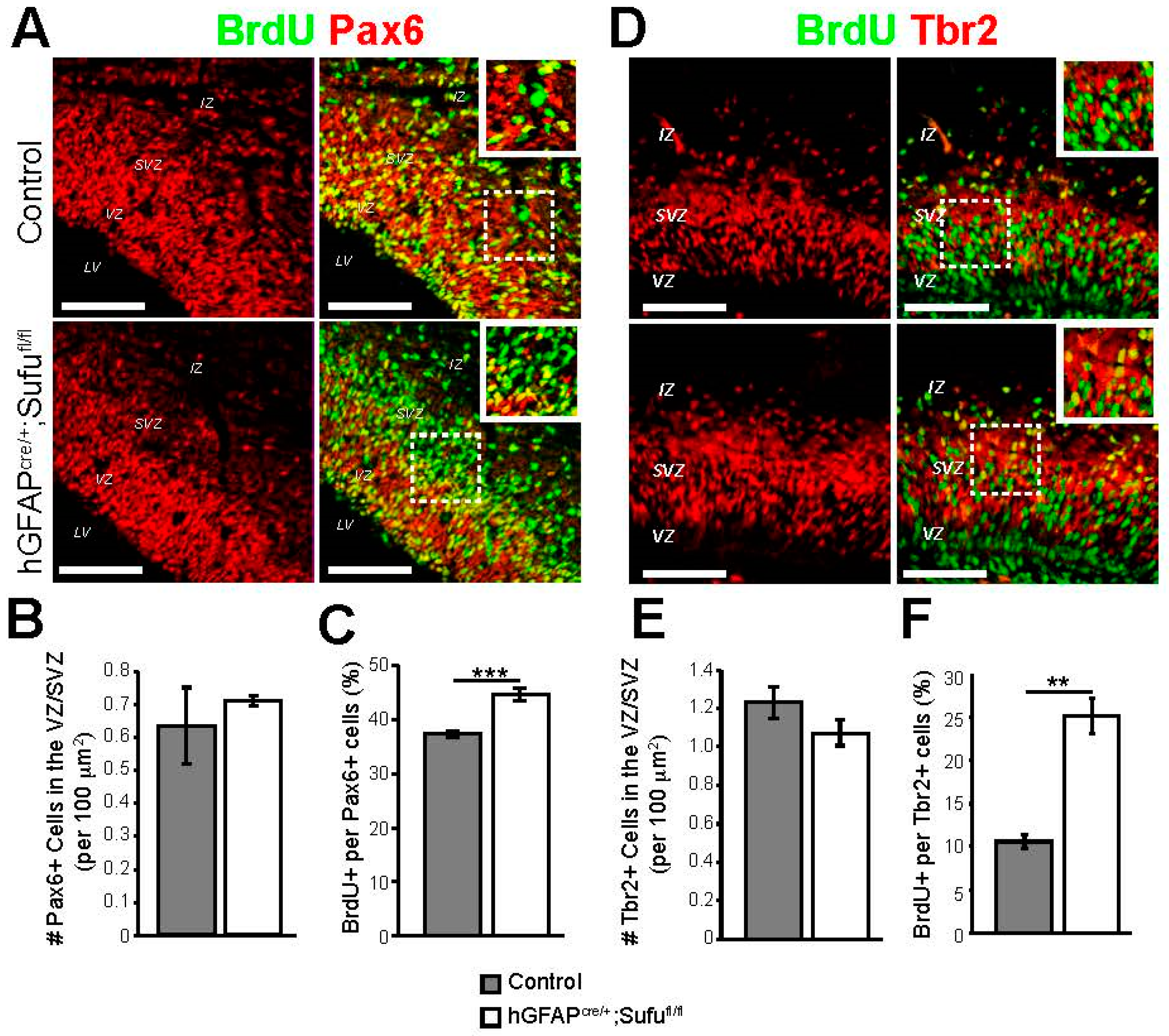
3.2. Abnormal Proliferation of Cortical Progenitors in the E16.5 Neocortex of hGFAPcre/+;Sufufl/fl Mice
3.3. Increase in Basally Dividing Progenitors in the E16.5 hGFAPcre/+;Sufufl/fl Neocortex
3.4. Reduced Levels of Gli3R in the E16.5 hGFAPcre/+;Sufufl/fl Neocortex
3.5. Activation of Shh Signaling in the E16.5 hGFAPcre/+;Sufufl/fl Neocortex
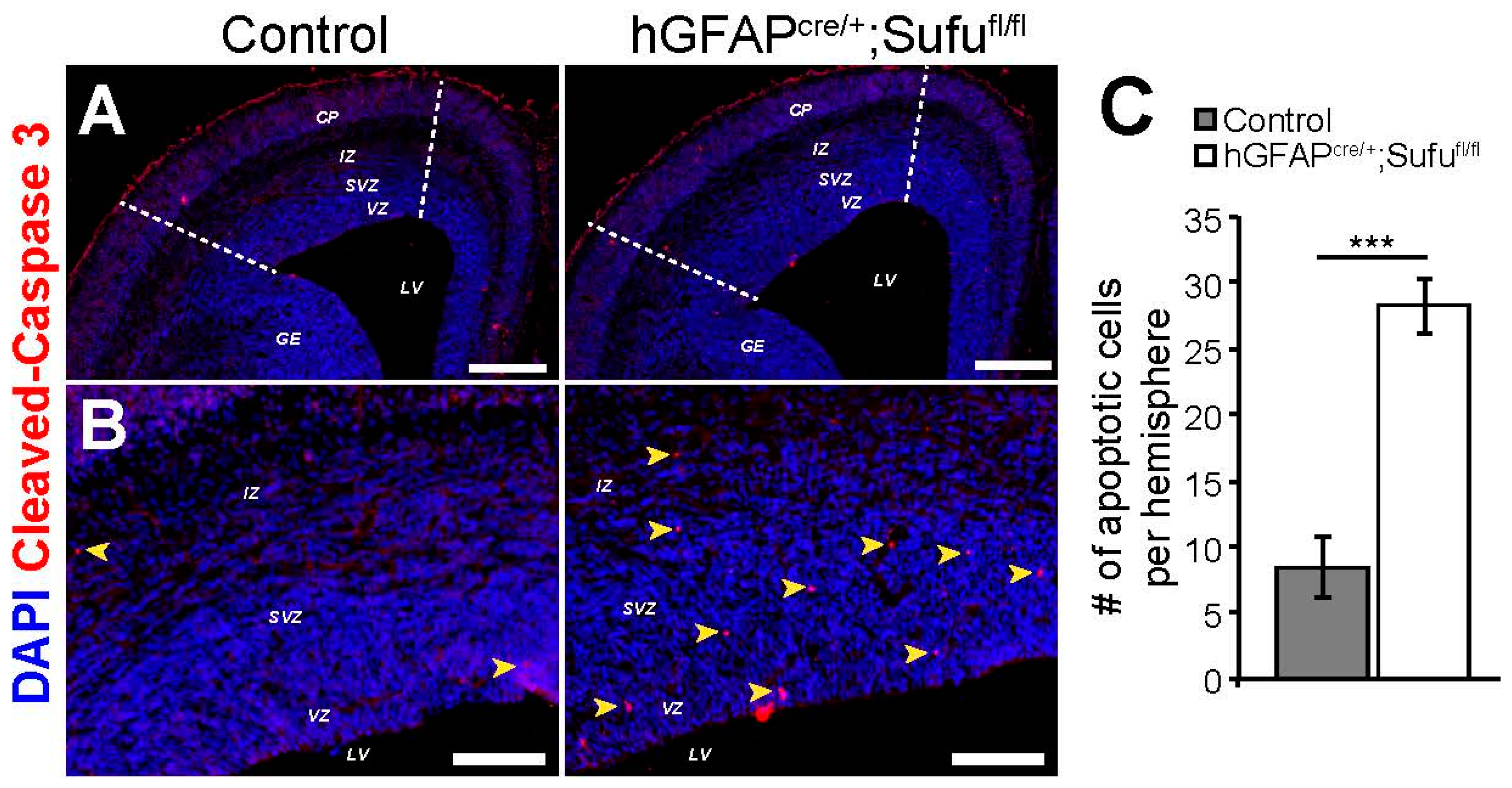
3.6. Increase in Cell Death in the E16.5 hGFAPcre/+;Sufufl/fl Neocortex
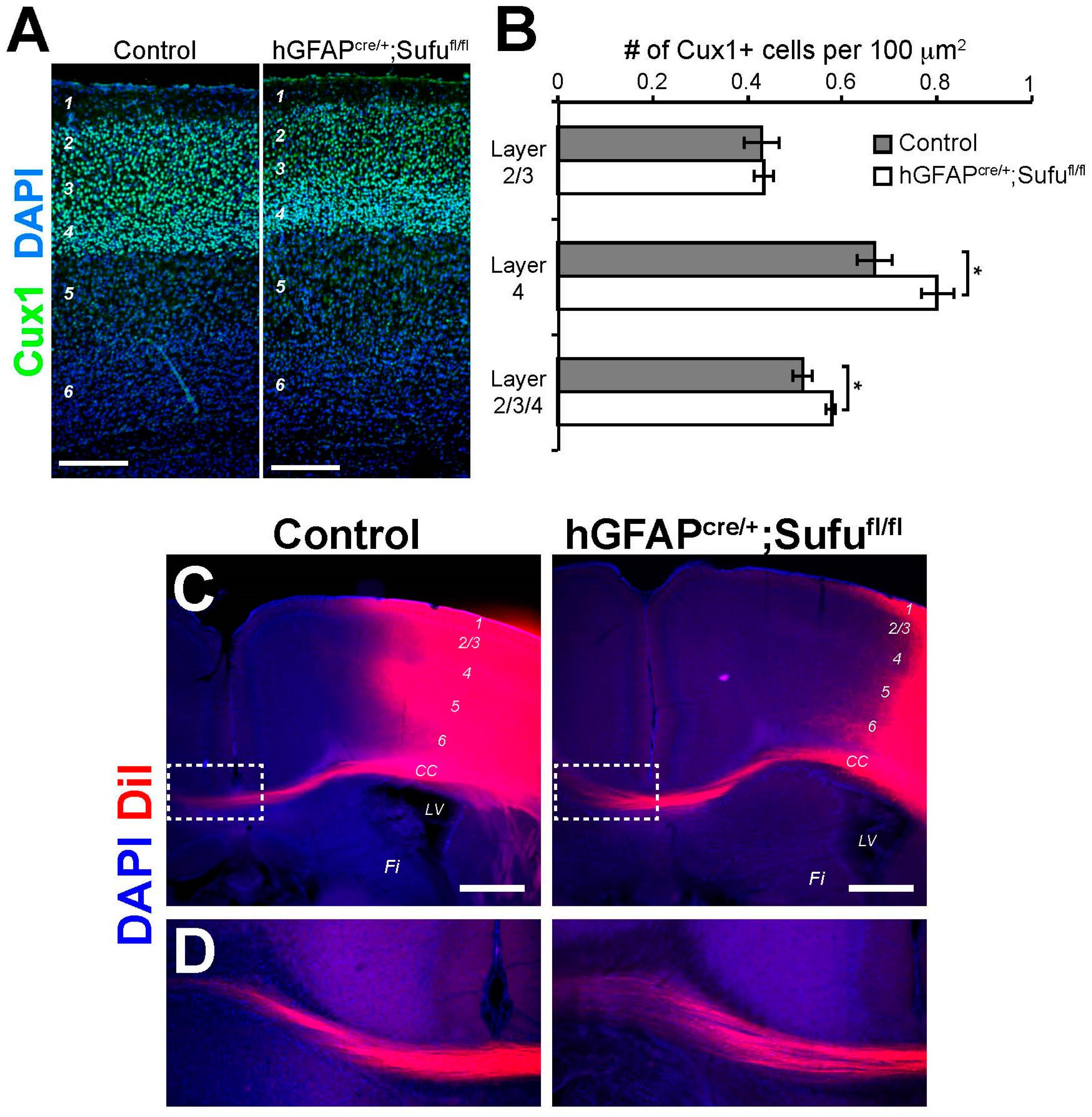
3.7. Increase in the Number of Cux1+ Upper Layer Neurons in the P7 hGFAPcre/+;Sufufl/fl Neocortex
3.8. Normal Formation of Callosal Projections in the P15 hGFAPcre/+;Sufufl/fl Neocortex
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Noctor, S.C.; Flint, A.C.; Weissman, T.A.; Dammerman, R.S.; Kriegstein, A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001, 409, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Noctor, S.C.; Martínez-Cerdeño, V.; Ivic, L.; Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004, 7, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Stancik, E.K.; Navarro-Quiroga, I.; Sellke, R.; Haydar, T.F. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J. Neurosci. 2010, 30, 7028–7036. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, T.; Pontious, A.; Englund, C.; Daza, R.A.M.; Bedogni, F.; Hodge, R.; Attardo, A.; Bell, C.; Huttner, W.B.; Hevner, R.F. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb. Cortex 2009, 19, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Sousa, V.H.; Fishell, G. Sonic hedgehog functions through dynamic changes in temporal competence in the developing forebrain. Curr. Opin. Genet. Dev. 2010, 20, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Yabut, O.R.; Fernandez, G.; Huynh, T.; Yoon, K.; Pleasure, S.J. Suppressor of Ffused is critical for maintenance of neuronal progenitor identity during corticogenesis. Cell Rep. 2015, 12, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ge, G.; Uchida, Y.; Luu, B.; Ahn, S. Gli3 is required for maintenance and fate specification of cortical progenitors. J. Neurosci. 2011, 31, 6440–6448. [Google Scholar] [CrossRef] [PubMed]
- Shikata, Y.; Okada, T.; Hashimoto, M.; Ellis, T.; Matsumaru, D.; Shiroishi, T.; Ogawa, M.; Wainwright, B.; Motoyama, J. Ptch1-mediated dosage-dependent action of Shh signaling regulates neural progenitor development at late gestational stages. Dev. Biol. 2011, 349, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Pozniak, C.D.; Langseth, A.J.; Dijkgraaf, G.J.P.; Choe, Y.; Werb, Z.; Pleasure, S.J. Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing Suppressor of Fused expression. Proc. Natl. Acad. Sci. USA 2010, 107, 21795–21800. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Heydeck, W.; Zeng, H.; Liu, A. Dual function of suppressor of fused in Hh pathway activation and mouse spinal cord patterning. Dev. Biol. 2012, 362, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Gill, P.S.; Rotin, L.; van Eede, M.; Henkelman, R.M.; Hui, C.-C.; Rosenblum, N.D. Suppressor of fused controls mid-hindbrain patterning and cerebellar morphogenesis via GLI3 repressor. J. Neurosci. 2011, 31, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chen, M.-H.; Yao, E.; Song, H.; Gacayan, R.; Hui, C.; Chuang, P.-T. Differential regulation of Gli proteins by Sufu in the lung affects PDGF signaling and myofibroblast development. Dev. Biol. 2014, 392, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, Y.; Wang, B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development 2010, 137, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Shahi, M.H.; Rey, J.A.; Castresana, J.S. The sonic hedgehog-GLI1 signaling pathway in brain tumor development. Expert Opin. Ther. Targets 2012, 16, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Komada, M.; Saitsu, H.; Kinboshi, M.; Miura, T.; Shiota, K.; Ishibashi, M. Hedgehog signaling is involved in development of the neocortex. Development 2008, 135, 2717–2727. [Google Scholar] [CrossRef] [PubMed]
- Pospisilik, J.A.; Schramek, D.; Schnidar, H.; Cronin, S.J.F.; Nehme, N.T.; Zhang, X.; Knauf, C.; Cani, P.D.; Aumayr, K.; Todoric, J.; et al. Drosophila genome-wide obesity screen reveals hedgehog as a determinant of brown versus white adipose cell fate. Cell 2010, 140, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Siegenthaler, J.A.; Ashique, A.M.; Zarbalis, K.; Patterson, K.P.; Hecht, J.H.; Kane, M.A.; Folias, A.E.; Choe, Y.; May, S.R.; Kume, T.; et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell 2009, 139, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Regard, J.B.; Malhotra, D.; Gvozdenovic-Jeremic, J.; Josey, M.; Chen, M.; Weinstein, L.S.; Lu, J.; Shore, E.M.; Kaplan, F.S.; Yang, Y. Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat. Med. 2013, 19, 1505–1512. [Google Scholar] [CrossRef] [PubMed]
- Lobo, S.; Wiczer, B.M.; Bernlohr, D.A. Functional analysis of long-chain acyl-CoA synthetase 1 in 3T3-L1 adipocytes. J. Biol. Chem. 2009, 284, 18347–18356. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, L.; Theis, M.; Alvarez-Maya, I.; Brenner, M.; Willecke, K.; Messing, A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 2001, 31, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Englund, C.; Fink, A.; Lau, C.; Pham, D.; Daza, R.A.M.; Bulfone, A.; Kowalczyk, T.; Hevner, R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005, 25, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Joyner, A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 2005, 437, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Fame, R.M.; MacDonald, J.L.; Macklis, J.D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011, 34, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Greig, L.C.; Woodworth, M.B.; Galazo, M.J.; Padmanabhan, H.; Macklis, J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013, 14, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hou, S.; Han, Y.-G. Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nat. Neurosci. 2016, 19, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Lien, W.-H.; Klezovitch, O.; Fernandez, T.E.; Delrow, J.; Vasioukhin, V. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science 2006, 311, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, N.; Sanchez, P.; Gitton, Y.; Palma, V.; Sun, T.; Beyna, M.; Weiner, H.; Ruiz i Altaba, A. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 2001, 128, 5201–5212. [Google Scholar] [PubMed]
- Humke, E.W.; Dorn, K.V.; Milenkovic, L.; Scott, M.P.; Rohatgi, R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010, 24, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Wilson, C.W.; Li, Y.-J.; Law, K.K.L.; Lu, C.-S.; Gacayan, R.; Zhang, X.; Hui, C.; Chuang, P.-T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009, 23, 1910–1928. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Du, W.; Chen, L.; Wang, G.; Cui, Y.; Liu, Y.; Dou, Z.; Wang, H.; Zhang, P.; et al. Suppressor of fused (Sufu) represses Gli1 transcription and nuclear accumulation, inhibits glioma cell proliferation, invasion and vasculogenic mimicry, improving glioma chemo-sensitivity and prognosis. Oncotarget 2014, 5, 11681–11694. [Google Scholar] [CrossRef] [PubMed]
- Kise, Y.; Morinaka, A.; Teglund, S.; Miki, H. Sufu recruits GSK3β for efficient processing of Gli3. Biochem. Biophys. Res. Commun. 2009, 387, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Makino, S.; Zhulyn, O.; Mo, R.; Puviindran, V.; Zhang, X.; Murata, T.; Fukumura, R.; Ishitsuka, Y.; Kotaki, H.; Matsumaru, D.; et al. T396I mutation of mouse Sufu reduces the stability and activity of Gli3 repressor. PLoS ONE 2015, 10, e0119455. [Google Scholar] [CrossRef] [PubMed]
- Petrova, R.; Garcia, A.D.R.; Joyner, A.L. Titration of GLI3 repressor activity by Sonic hedgehog signaling is critical for maintaining multiple adult neural stem cell and astrocyte functions. J. Neurosci. 2013, 33, 17490–17505. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.K.; Fuentealba, L.C.; Shah, J.K.; Lindquist, R.A.; Ihrie, R.A.; Guinto, C.D.; Rodas-Rodriguez, J.L.; Alvarez-Buylla, A. A Dorsal SHH-Dependent Domain in the V-SVZ Produces Large Numbers of Oligodendroglial Lineage Cells in the Postnatal Brain. Stem Cell Rep. 2015, 5, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Min, T.H.; Kriebel, M.; Hou, S.; Pera, E.M. The dual regulator Sufu integrates Hedgehog and Wnt signals in the early Xenopus embryo. Dev. Biol. 2011, 358, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Poon, R.; Zhang, X.; Cheah, A.; Ding, Q.; Hui, C.C.; Alman, B. Suppressor of fused negatively regulates beta-catenin signaling. J. Biol. Chem. 2001, 276, 40113–40119. [Google Scholar] [CrossRef] [PubMed]
- Harrison-Uy, S.J.; Pleasure, S.J. Wnt signaling and forebrain development. Cold Spring Harb. Perspect. Biol. 2012, 4, a008094. [Google Scholar] [CrossRef] [PubMed]
- Tyler, W.A.; Medalla, M.; Guillamon-Vivancos, T.; Luebke, J.I.; Haydar, T.F. Neural precursor lineages specify distinct neocortical pyramidal neuron types. J. Neurosci. 2015, 35, 6142–6152. [Google Scholar] [CrossRef] [PubMed]
- Mihalas, A.B.; Elsen, G.E.; Bedogni, F.; Daza, R.A.M.; Ramos-Laguna, K.A.; Arnold, S.J.; Hevner, R.F. Intermediate Progenitor Cohorts Differentially Generate Cortical Layers and Require Tbr2 for Timely Acquisition of Neuronal Subtype Identity. Cell Rep. 2016, 16, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Courchesne, E.; Mouton, P.R.; Calhoun, M.E.; Semendeferi, K.; Ahrens-Barbeau, C.; Hallet, M.J.; Barnes, C.C.; Pierce, K. Neuron number and size in prefrontal cortex of children with autism. JAMA 2011, 306, 2001–2010. [Google Scholar] [CrossRef] [PubMed]
- Casanova, M.F.; van Kooten, I.A.J.; Switala, A.E.; van Engeland, H.; Heinsen, H.; Steinbusch, H.W.M.; Hof, P.R.; Trippe, J.; Stone, J.; Schmitz, C. Minicolumnar abnormalities in autism. Acta Neuropathol. 2006, 112, 287–303. [Google Scholar] [CrossRef] [PubMed]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yabut, O.R.; Ng, H.X.; Fernandez, G.; Yoon, K.; Kuhn, J.; Pleasure, S.J. Loss of Suppressor of Fused in Mid-Corticogenesis Leads to the Expansion of Intermediate Progenitors. J. Dev. Biol. 2016, 4, 29. https://doi.org/10.3390/jdb4040029
Yabut OR, Ng HX, Fernandez G, Yoon K, Kuhn J, Pleasure SJ. Loss of Suppressor of Fused in Mid-Corticogenesis Leads to the Expansion of Intermediate Progenitors. Journal of Developmental Biology. 2016; 4(4):29. https://doi.org/10.3390/jdb4040029
Chicago/Turabian StyleYabut, Odessa R., Hui Xuan Ng, Gloria Fernandez, Keejung Yoon, Jeremy Kuhn, and Samuel J. Pleasure. 2016. "Loss of Suppressor of Fused in Mid-Corticogenesis Leads to the Expansion of Intermediate Progenitors" Journal of Developmental Biology 4, no. 4: 29. https://doi.org/10.3390/jdb4040029




