Differential Cellular Responses to Hedgehog Signalling in Vertebrates—What is the Role of Competence?
Abstract
:1. Introduction
- Cellular responses may depend on cell-intrinsic factors that reflect the context of the responding cell, its location and developmental history (Figure 1B). This contextual ability to respond to a signal brings us back to the idea of cellular competence: cells that respond differently to a given signal display differential competence for this signal.
2. A Brief Outline of the Hedgehog Pathway
3. Roles of Hedgehog Signalling
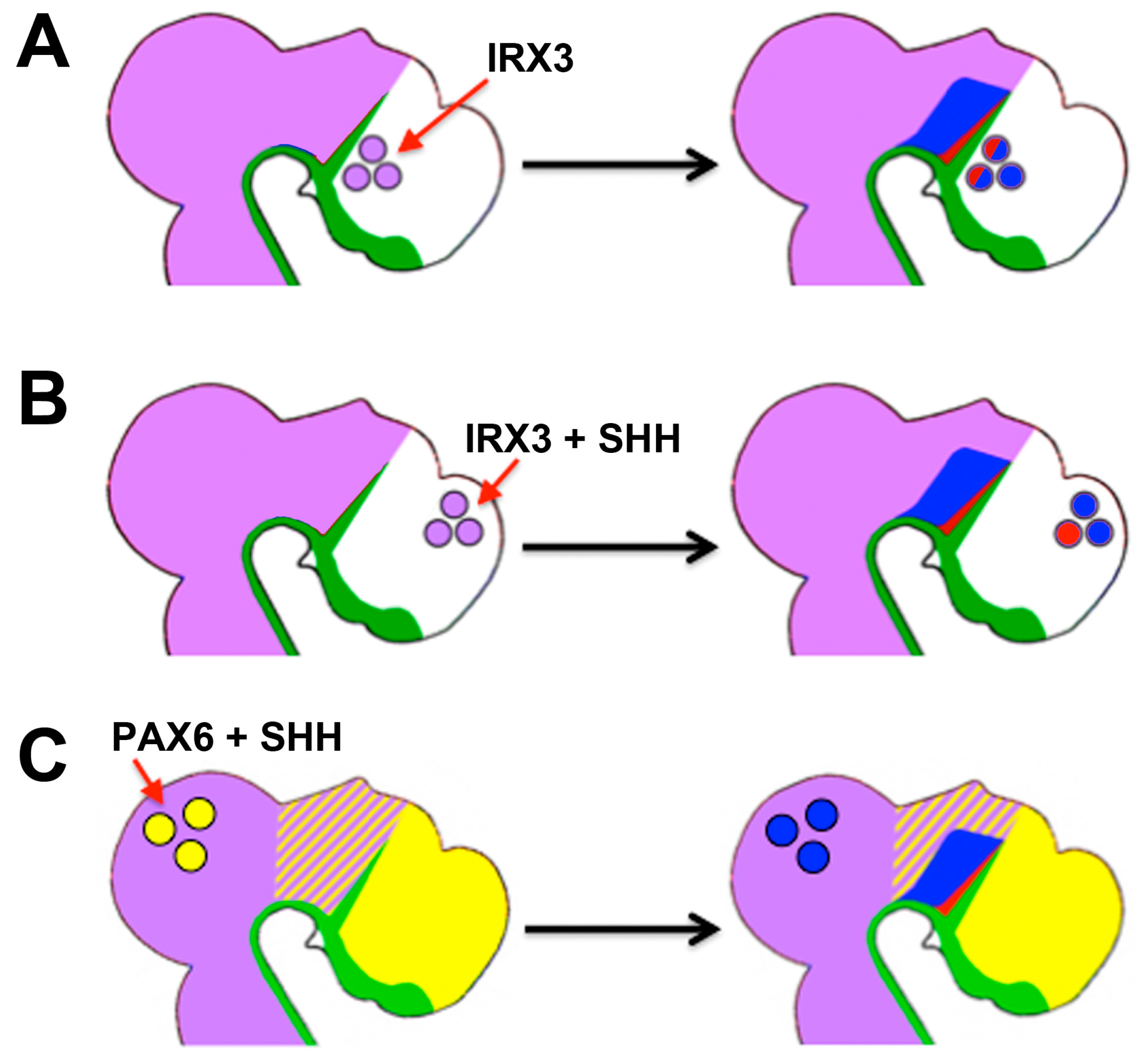
4. Differential Competence for Hedgehog
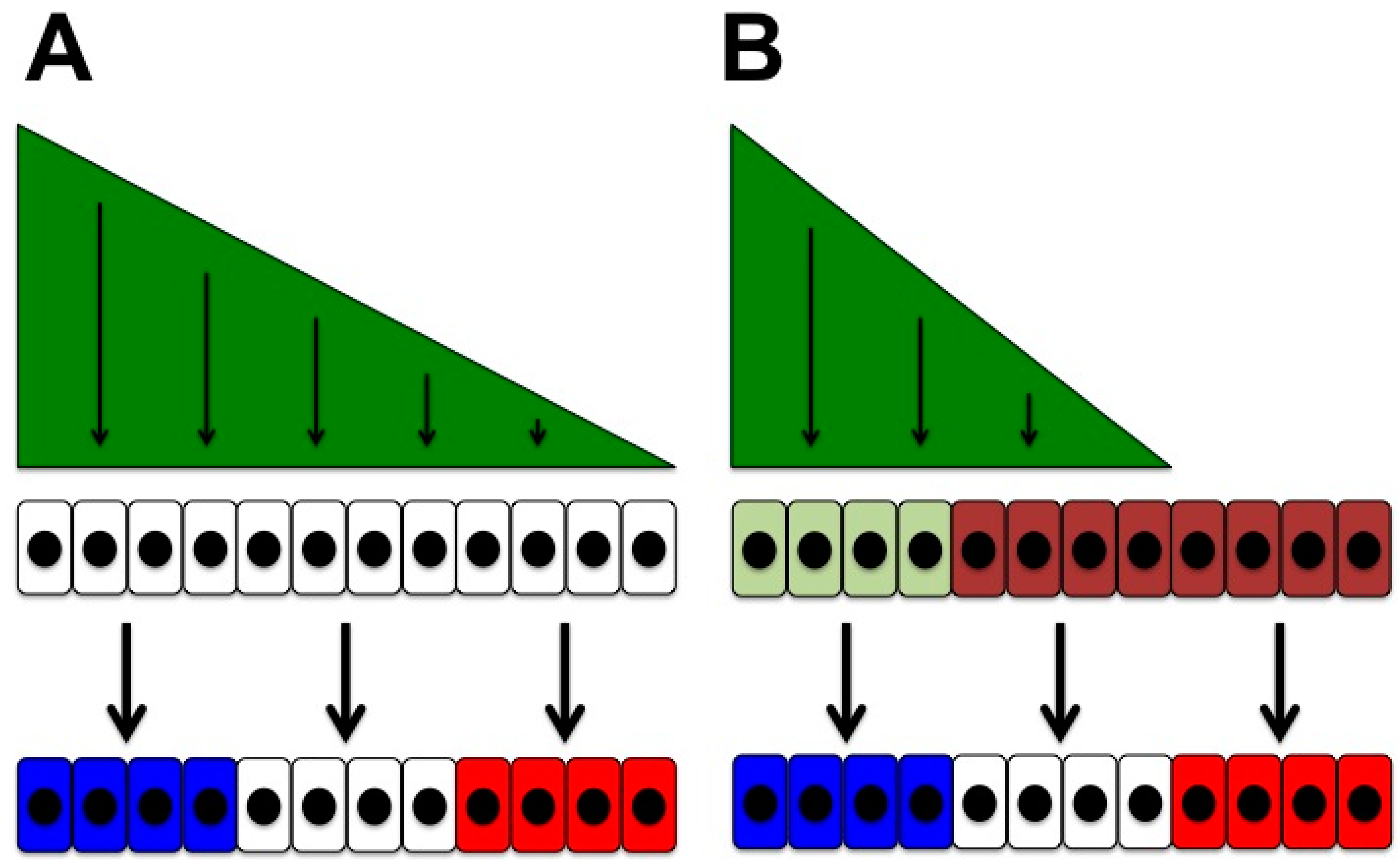
4.1. Temporal Changes in Competence for Hedgehog Signalling
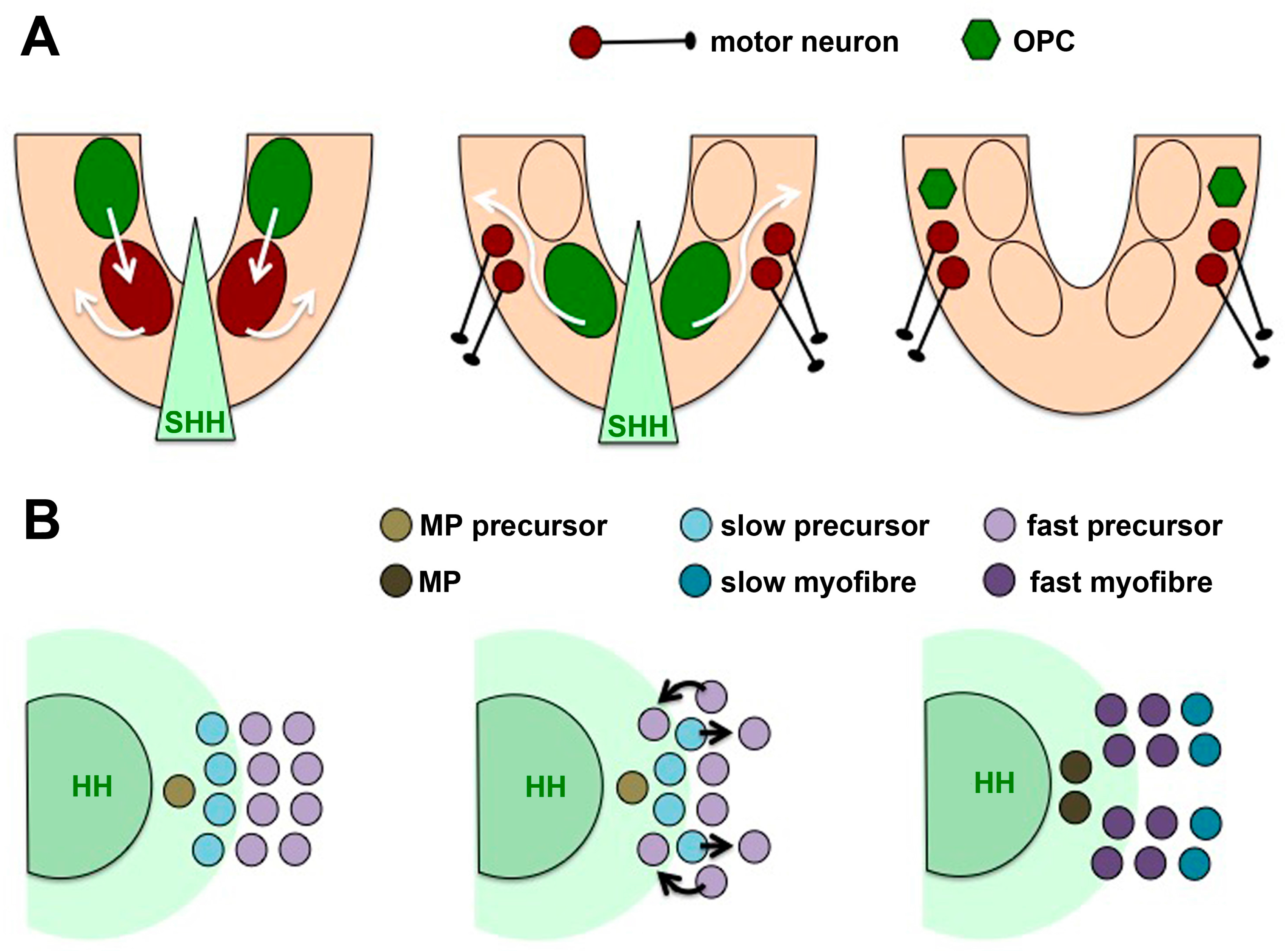
4.2. Cells Movements May Accompany Temporal Competence Changes
4.3. Other Signalling Pathways Modulate Cellular Responses to Hedgehog Signalling
4.4. Receptor Switching Can Change the Competence for Hedgehog Signalling
4.5. Differential Competence for HH Signalling in Different Tissue Types
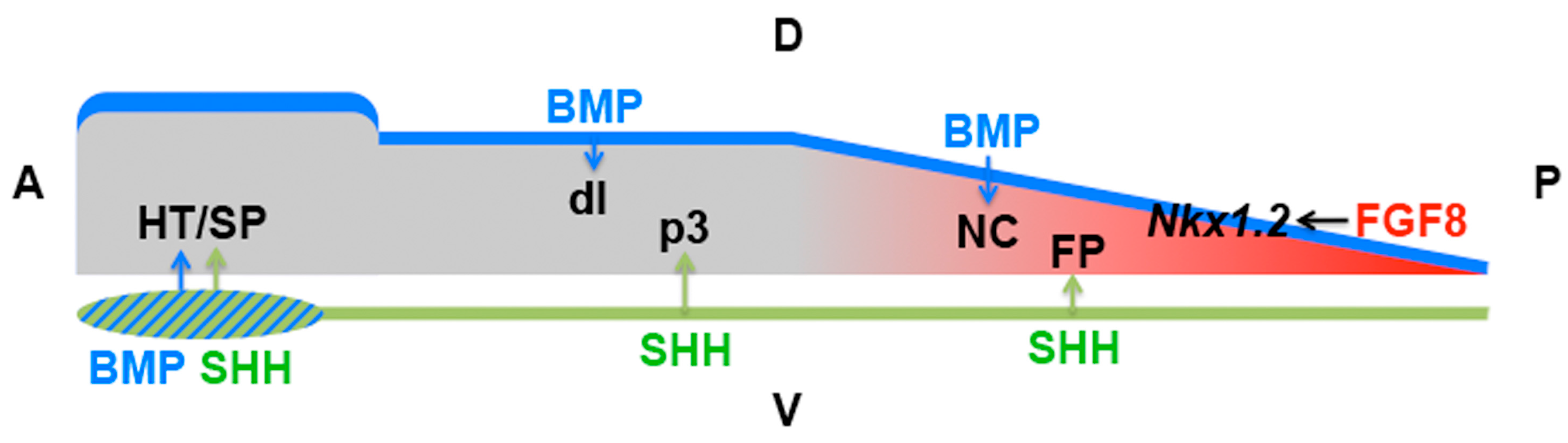
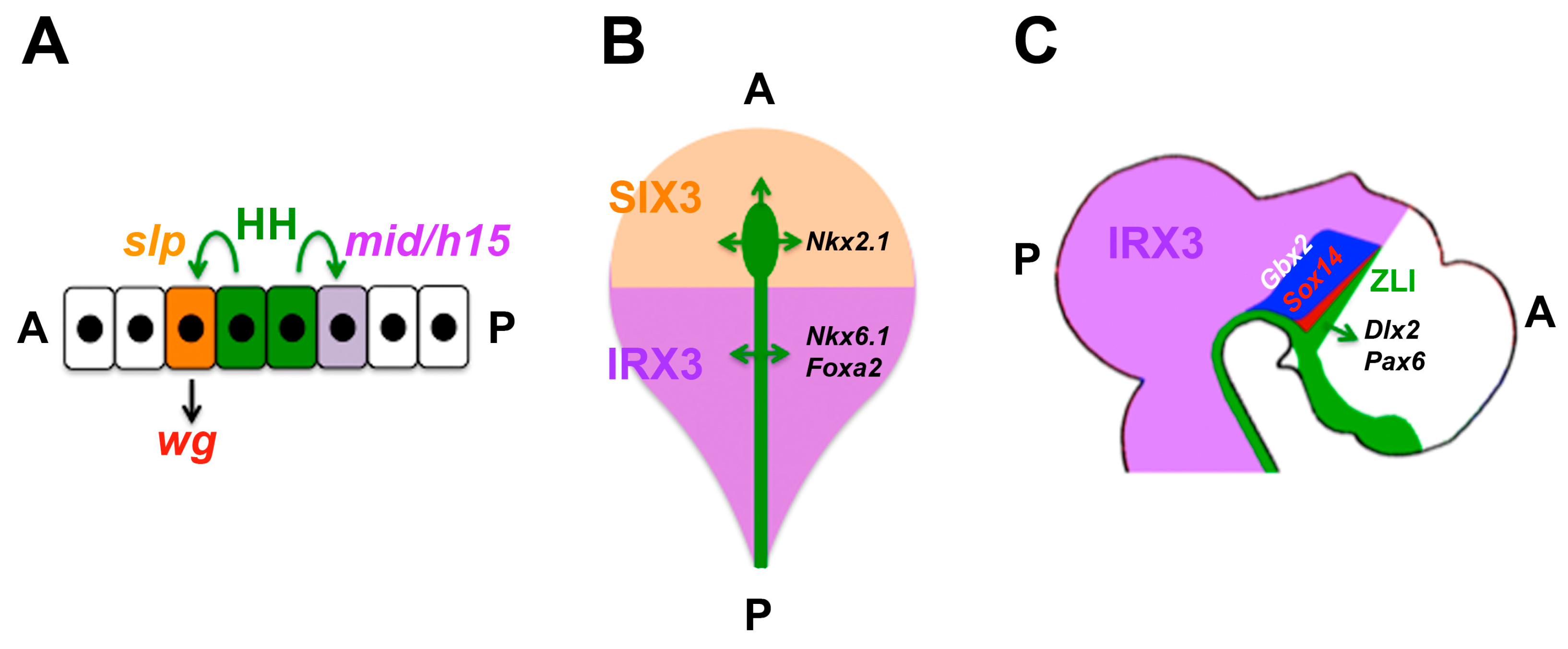
4.6. Domains of Differential Competence for HH Signalling within a Tissue
5. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Spemann, H. Über Correlationen in der Entwicklung des Auges. Verhand. Anat. Ges. 1901, 15, 61–79. [Google Scholar]
- Waddington, C.H. Organisers and Genes; Cambridge University Press: Cambridge, UK, 1940. [Google Scholar]
- Briscoe, J.; Small, S. Morphogen rules: Design principles of gradient-mediated embryo patterning. Development 2015, 142, 3996–4009. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; Nakano, Y.; Seger, C. Mechanisms and functions of hedgehog signalling across the metazoa. Nat. Rev. Genet. 2011, 12, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Taipale, J.; Cooper, M.K.; Maiti, T.; Beachy, P.A. Patched acts catalytically to suppress the activity of Smoothened. Nature 2002, 418, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Tukachinsky, H.; Petrov, K.; Watanabe, M.; Salic, A. Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. USA 2016, 113, E5866–E5875. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, M.F.; Spek, C.A.; Zivkovic, D.; van de Water, S.; Rezaee, F.; Peppelenbosch, M.P. Repression of smoothened by patched-dependent (pro-)vitamin D3 secretion. PLoS Biol. 2006, 4, e232. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Weber, S.; Dittmann, K.; Adamski, J.; Hahn, H.; Uhmann, A. A Functional and Putative Physiological Role of Calcitriol in Patched1/Smoothened Interaction. J. Biol. Chem. 2015, 290, 19614–19628. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, A.C.; Roberts, B.; Kwong, L.; Bijlsma, M.F.; Roelink, H. Ptch2 mediates the Shh response in Ptch1-/- cells. Development 2014, 141, 3331–3339. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Buttitta, L.; Fan, C.M. Evidence that the WNT-inducible growth arrest-specific gene 1 encodes an antagonist of sonic hedgehog signaling in the somite. Proc. Natl. Acad. Sci. USA 2001, 98, 11347–11352. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Lum, L.; Beachy, P. The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 2006, 125, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Allen, B.L.; Tenzen, T.; McMahon, A.P. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007, 21, 1244–1257. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, D.C.; Fan, C.M. Gas1 extends the range of Hedgehog action by facilitating its signaling. Genes Dev. 2007, 21, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Tenzen, T.; Allen, B.L.; Cole, F.; Kang, J.S.; Krauss, R.S.; McMahon, A.P. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev. Cell 2006, 10, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Chuang, P.T.; McMahon, A.P. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 1999, 397, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Kwong, L.; Bijlsma, M.F.; Roelink, H. Shh-mediated degradation of Hhip allows cell autonomous and non-cell autonomous Shh signalling. Nat. Commun. 2014, 5, 4849. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tong, C.; Jiang, J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature 2007, 450, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Corbit, K.C.; Aanstad, P.; Singla, V.; Norman, A.R.; Stainier, D.Y.; Reiter, J.F. Vertebrate Smoothened functions at the primary cilium. Nature 2005, 437, 1018–1021. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Z.; Walsh, C.T.; McMahon, A.P. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc. Natl. Acad. Sci. USA 2009, 106, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Goetz, S.C.; Anderson, K.V. The primary cilium: A signalling centre during vertebrate development. Nat. Rev. Genet. 2010, 11, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Haycraft, C.J.; Banizs, B.; Aydin-Son, Y.; Zhang, Q.; Michaud, E.J.; Yoder, B.K. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005, 1, e53. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, R.J.; Tobin, J.L.; Beales, P.L. Modeling ciliopathies: Primary cilia in development and disease. Curr. Top. Dev. Biol. 2008, 84, 249–310. [Google Scholar] [PubMed]
- Briscoe, J.; Thérond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Robbins, D.J.; Nybakken, K.E.; Kobayashi, R.; Sisson, J.C.; Bishop, J.M.; Thérond, P.P. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 1997, 90, 225–234. [Google Scholar] [CrossRef]
- Sisson, J.C.; Ho, K.S.; Suyama, K.; Scott, M.P. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell 1997, 90, 235–245. [Google Scholar] [CrossRef]
- Ingham, P.W.; McMahon, A.P. Hedgehog signalling: Kif7 is not that fishy after all. Curr. Biol. 2009, 19, R729–R731. [Google Scholar] [CrossRef] [PubMed]
- Charron, F.; Tessier-Lavigne, M. Novel brain wiring functions for classical morphogens: A role as graded positional cues in axon guidance. Development 2005, 132, 2251–2262. [Google Scholar] [CrossRef] [PubMed]
- Varjosalo, M.; Björklund, M.; Cheng, F.; Syvänen, H.; Kivioja, T.; Kilpinen, S.; Sun, Z.; Kallioniemi, O.; Stunnenberg, H.G.; He, W.W.; et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell 2008, 133, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Yam, P.T.; Langlois, S.D.; Morin, S.; Charron, F. Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron 2009, 62, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Belgacem, Y.H.; Borodinsky, L.N. Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc. Natl. Acad. Sci. USA 2011, 108, 4482–4487. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, P.; Xiao, L.; Kazanietz, M.G.; Riobo, N.A. Hedgehog proteins activate pro-angiogenic responses in endothelial cells through non-canonical signaling pathways. Cell Cycle 2010, 9, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Renault, M.A.; Roncalli, J.; Tongers, J.; Thorne, T.; Klyachko, E.; Misener, S.; Volpert, O.V.; Mehta, S.; Burg, A.; Luedemann, C.; et al. Sonic hedgehog induces angiogenesis via Rho kinase-dependent signaling in endothelial cells. J. Mol. Cell. Cardiol. 2010, 49, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Polizio, A.H.; Chinchilla, P.; Chen, X.; Manning, D.R.; Riobo, N.A. Sonic hedgehog activates the GTPases Rac1 and RhoA in a Gli-independent manner through coupling of smoothened to Gi proteins. Sci. Signal. 2011, 4, pt7. [Google Scholar] [CrossRef] [PubMed]
- Polizio, A.H.; Chinchilla, P.; Chen, X.; Kim, S.; Manning, D.R.; Riobo, N.A. Heterotrimeric Gi proteins link Hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J. Biol. Chem. 2011, 286, 19589–19596. [Google Scholar] [CrossRef] [PubMed]
- Thibert, C.; Teillet, M.A.; Lapointe, F.; Mazelin, L.; Le Douarin, N.M.; Mehlen, P. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science 2003, 301, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Delloye-Bourgeois, C.; Gibert, B.; Rama, N.; Delcros, J.G.; Gadot, N.; Scoazec, J.Y.; Krauss, R.; Bernet, A.; Mehlen, P. Sonic hedgehog promotes tumor cell survival by inhibiting CDON pro-apoptotic activity. PLoS Biol. 2013, 11, e1001623. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.E.; Scott, M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005, 6, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Robbins, D.J.; Fei, D.L.; Riobo, N.A. The Hedgehog signal transduction network. Sci. Signal. 2012, 5, re6. [Google Scholar] [CrossRef] [PubMed]
- Nüsslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Forbes, A.J.; Nakano, Y.; Taylor, A.M.; Ingham, P.W. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev. Suppl. 1993, 115–124. [Google Scholar]
- Basler, K.; Struhl, G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature 1994, 368, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Blair, S.S. Developmental biology. Hedgehog digs up an old friend. Nature 1995, 373, 656–657. [Google Scholar] [CrossRef] [PubMed]
- Echelard, Y.; Epstein, D.J.; St-Jacques, B.; Shen, L.; Mohler, J.; McMahon, J.A.; McMahon, A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 1993, 75, 1417–1430. [Google Scholar] [CrossRef]
- Placzek, M.; Briscoe, J. The floor plate: Multiple cells, multiple signals. Nat. Rev. Neurosci. 2005, 6, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Fuccillo, M.; Joyner, A.L.; Fishell, G. Morphogen to mitogen: The multiple roles of hedgehog signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006, 7, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Lupo, G.; Harris, W.A.; Lewis, K.E. Mechanisms of ventral patterning in the vertebrate nervous system. Nat. Rev. Neurosci. 2006, 7, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Dessaud, E.; McMahon, A.P.; Briscoe, J. Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen-regulated transcriptional network. Development 2008, 135, 2489–2503. [Google Scholar] [CrossRef] [PubMed]
- Kiecker, C.; Lumsden, A. The role of organizers in patterning the nervous system. Annu. Rev. Neurosci. 2012, 35, 347–367. [Google Scholar] [CrossRef] [PubMed]
- Blaess, S.; Szabo, N.; Haddad-Tovolli, R.; Zhou, X.; Alvarez-Bolado, G. Sonic hedgehog signaling in the development of the mouse hypothalamus. Front. Neuroanat. 2014, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Blaess, S.; Ang, S.L. Genetic control of midbrain dopaminergic neuron development. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Roessler, E.; Muenke, M. The molecular genetics of holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010, 154C, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, A. Next generation limb development and evolution: Old questions, new perspectives. Development 2015, 142, 3810–3820. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.E.; Roessler, E.; Muenke, M. Human developmental disorders and the Sonic hedgehog pathway. Mol. Med. Today 1998, 4, 343–349. [Google Scholar] [CrossRef]
- Lettice, L.A.; Hill, R.E. Preaxial polydactyly: A model for defective long-range regulation in congenital abnormalities. Curr. Opin. Genet. Dev. 2005, 15, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.M.; Tessier-Lavigne, M. Patterning of mammalian somites by surface ectoderm and notochord: Evidence for sclerotome induction by a hedgehog homolog. Cell 1994, 79, 1175–1186. [Google Scholar] [CrossRef]
- Johnson, R.L.; Laufer, E.; Riddle, R.D.; Tabin, C. Ectopic expression of Sonic hedgehog alters dorsal-ventral patterning of somites. Cell 1994, 79, 1165–1173. [Google Scholar] [CrossRef]
- Karlstrom, R.O.; Talbot, W.S.; Schier, A.F. Comparative synteny cloning of zebrafish you-too: Mutations in the hedgehog target gli2 affect ventral forebrain patterning. Genes Dev. 1999, 13, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Hebrok, M. Intercellular signals regulating pancreas development and function. Genes Dev. 2001, 15, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Jackson, H.E.; Ingham, P.W. Control of muscle fibre-type diversity during embryonic development: The zebrafish paradigm. Mech. Dev. 2013, 130, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Seidel, K.; Salcedo, E.; Ahn, C.; de Sauvage, F.J.; Klein, O.D.; Barlow, L.A. Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development 2014, 141, 2993–3002. [Google Scholar] [CrossRef] [PubMed]
- St-Jacques, B.; Dassule, H.R.; Karavanova, I.; Botchkarev, V.A.; Li, J.; Danielian, P.S.; McMahon, J.A.; Lewis, P.M.; Paus, R.; McMahon, A.P. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 1998, 8, 1058–1068. [Google Scholar] [CrossRef]
- Balic, A.; Thesleff, I. Tissue interactions regulating tooth development and renewal. Curr. Top. Dev. Biol. 2015, 115, 157–186. [Google Scholar] [PubMed]
- Bitgood, M.J.; Shen, L.; McMahon, A.P. Sertoli cell signaling by Desert hedgehog regulates the male germline. Curr. Biol. 1996, 6, 298–304. [Google Scholar] [CrossRef]
- Vortkamp, A.; Lee, K.; Lanske, B.; Segre, G.V.; Kronenberg, H.M.; Tabin, C.J. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 1996, 273, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Dahmane, N.; Ruiz i Altaba, A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999, 126, 3089–3100. [Google Scholar] [PubMed]
- Wallace, V.A. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 1999, 9, 445–448. [Google Scholar] [CrossRef]
- Wechsler-Reya, R.J.; Scott, M.P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef]
- Britto, J.; Tannahill, D.; Keynes, R. A critical role for sonic hedgehog signaling in the early expansion of the developing brain. Nat. Neurosci. 2002, 5, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; McMahon, A.P. A sonic hedgehog-dependent signaling relay regulates growth of diencephalic and mesencephalic primordia in the early mouse embryo. Development 2002, 129, 4807–4819. [Google Scholar] [PubMed]
- Kenney, A.M.; Rowitch, D.H. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 2000, 20, 9055–9067. [Google Scholar] [CrossRef] [PubMed]
- Kenney, A.M.; Cole, M.D.; Rowitch, D.H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 2003, 130, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.G.; Grasfeder, L.L.; Carroll, A.L.; Kaiser, C.; Gillingham, C.L.; Lin, S.M.; Wickramasinghe, R.; Scott, M.P.; Wechsler-Reya, R.J. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc. Natl. Acad. Sci. USA 2003, 100, 7331–7336. [Google Scholar] [CrossRef] [PubMed]
- Barnes, E.A.; Kong, M.; Ollendorff, V.; Donoghue, D.J. Patched1 interacts with Cyclin b1 to regulate cell cycle progression. EMBO J. 2001, 20, 2214–2223. [Google Scholar] [CrossRef] [PubMed]
- Crosnier, C.; Stamataki, D.; Lewis, J. Organizing cell renewal in the intestine: Stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006, 7, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Rowitch, D.H.; S-Jacques, B.; Lee, S.M.; Flax, J.D.; Snyder, E.Y.; McMahon, A.P. Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J. Neurosci. 1999, 19, 8954–8965. [Google Scholar] [PubMed]
- Lai, K.; Kaspar, B.K.; Gage, F.H.; Schaffer, D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003, 6, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Machold, R.; Hayashi, S.; Rutlin, M.; Muzumdar, M.D.; Nery, S.; Corbin, J.G.; Gritli-Linde, A.; Dellovade, T.; Porter, J.A.; Rubin, L.L.; et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron 2003, 39, 937–950. [Google Scholar] [CrossRef]
- Ahn, S.; Joyner, A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 2005, 437, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Aoto, K.; Shikata, Y.; Higashiyama, D.; Shiota, K.; Motoyama, J. Fetal ethanol exposure activates protein kinase a and impairs shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Res. A Clin. Mol. Teratol. 2008, 82, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Amankulor, N.M.; Hambardzumyan, D.; Pyonteck, S.M.; Becher, O.J.; Joyce, J.A.; Holland, E.C. Sonic hedgehog pathway activation is induced by acute brain injury and regulated by injury-related inflammation. J. Neurosci. 2009, 29, 10299–10308. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.I.; Shtaya, A.B.; Zaben, M.J.; Owens, E.V.; Kiecker, C.; Gray, W.P. Endogenous GFAP-positive neural stem/progenitor cells in the postnatal mouse cortex are activated following traumatic brain injury. J. Neurotrauma 2012, 29, 828–842. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.; Lee, J.; Guo, N.; Kim, J.; Lim, A.; Qu, L.; Mysorekar, I.U.; Beachy, P.A. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature 2011, 472, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Hahn, H.; Wicking, C.; Zaphiropoulous, P.G.; Gailani, M.R.; Shanley, S.; Chidambaram, A.; Vorechovsky, I.; Holmberg, E.; Unden, A.B.; Gillies, S.; et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 1996, 85, 841–851. [Google Scholar] [CrossRef]
- Johnson, R.L.; Rothman, A.L.; Xie, J.; Goodrich, L.V.; Bare, J.W.; Bonifas, J.M.; Quinn, A.G.; Myers, R.M.; Cox, D.R.; Epstein, E.H., Jr.; et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 1996, 272, 1668–1671. [Google Scholar] [CrossRef] [PubMed]
- Gajjar, A.J.; Robinson, G.W. Medulloblastoma-translating discoveries from the bench to the bedside. Nat. Rev. Clin. Oncol. 2014, 11, 714–722. [Google Scholar] [CrossRef] [PubMed]
- Aavikko, M.; Li, S.P.; Saarinen, S.; Alhopuro, P.; Kaasinen, E.; Morgunova, E.; Li, Y.; Vesanen, K.; Smith, M.J.; Evans, D.G.; et al. Loss of SUFU function in familial multiple meningioma. Am. J. Hum. Genet. 2012, 91, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Kijima, C.; Miyashita, T.; Suzuki, M.; Oka, H.; Fujii, K. Two cases of nevoid basal cell carcinoma syndrome associated with meningioma caused by a PTCH1 or SUFU germline mutation. Fam. Cancer 2012, 11, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.E.; Erson-Omay, E.Z.; Serin, A.; Yin, J.; Cotney, J.; Ozduman, K.; Avsar, T.; Li, J.; Murray, P.B.; Henegariu, O.; et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science 2013, 339, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Tostar, U.; Malm, C.J.; Meis-Kindblom, J.M.; Kindblom, L.G.; Toftgard, R.; Unden, A.B. Deregulation of the hedgehog signalling pathway: A possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J. Pathol. 2006, 208, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Amakye, D.; Jagani, Z.; Dorsch, M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013, 19, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Xie, J. Non-Canonical Hh Signaling in Cancer-Current Understanding and Future Directions. Cancers (Basel) 2015, 7, 1684–1698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, J.; McMahon, A.P. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched1 and Hhip1. Development 2005, 132, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Kicheva, A.; Bollenbach, T.; Ribeiro, A.; Valle, H.P.; Lovell-Badge, R.; Episkopou, V.; Briscoe, J. Coordination of progenitor specification and growth in mouse and chick spinal cord. Science 2014, 345, 1254927. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Anderson, D.J. The bHLH transcription factors Olig2 and Olig1 couple neuronal and glial subtype specification. Cell 2002, 109, 61–73. [Google Scholar] [CrossRef]
- Hochstim, C.; Deneen, B.; Lukaszewicz, A.; Zhou, Q.; Anderson, D.J. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell 2008, 133, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; McGlynn, S.; Matise, M.P. Floor plate-derived sonic hedgehog regulates glial and ependymal cell fates in the developing spinal cord. Development 2013, 140, 1594–1604. [Google Scholar] [CrossRef] [PubMed]
- Sousa, V.H.; Fishell, G. Sonic hedgehog functions through dynamic changes in temporal competence in the developing forebrain. Curr. Opin. Genet. Dev. 2010, 20, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Nakamura, E.; Nguyen, M.T.; Bao, X.; Akiyama, H.; Mackem, S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell 2008, 14, 624–632. [Google Scholar] [CrossRef] [PubMed]
- Balaskas, N.; Ribeiro, A.; Panovska, J.; Dessaud, E.; Sasai, N.; Page, K.M.; Briscoe, J.; Ribes, V. Gene regulatory logic for reading the Sonic hedgehog signaling gradient in the vertebrate neural tube. Cell 2012, 148, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Kicheva, A.; Ribeiro, A.; Blassberg, R.; Page, K.M.; Barnes, C.P.; Briscoe, J. Ptch1 and Gli regulate Shh signalling dynamics via multiple mechanisms. Nat. Commun. 2015, 6, 6709. [Google Scholar] [CrossRef] [PubMed]
- Sussel, L.; Marín, O.; Kimura, S.; Rubenstein, J.L. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: Evidence for a transformation of the pallidum into the striatum. Development 1999, 126, 3359–3370. [Google Scholar] [PubMed]
- Cayuso, J.; Ulloa, F.; Cox, B.; Briscoe, J.; Martí, E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development 2006, 133, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Mehta, A.; Richardson, J.S.; Appel, B. Olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev. Biol. 2002, 248, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Zannino, D.A.; Appel, B. Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. J. Neurosci. 2009, 29, 2322–2333. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Shin, J.; Appel, B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development 2004, 131, 5959–5969. [Google Scholar] [CrossRef] [PubMed]
- Ravanelli, A.M.; Appel, B. Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev. 2015, 29, 2504–2515. [Google Scholar] [CrossRef] [PubMed]
- Blagden, C.S.; Currie, P.D.; Ingham, P.W.; Hughes, S.M. Notochord induction of zebrafish slow muscle mediated by Sonic hedgehog. Genes Dev. 1997, 11, 2163–2175. [Google Scholar] [CrossRef] [PubMed]
- Wolff, C.; Roy, S.; Ingham, P.W. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr. Biol. 2003, 13, 1169–1181. [Google Scholar] [CrossRef]
- Baxendale, S.; Davison, C.; Muxworthy, C.; Wolff, C.; Ingham, P.W.; Roy, S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 2004, 36, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Ribes, V.; Balaskas, N.; Sasai, N.; Cruz, C.; Dessaud, E.; Cayuso, J.; Tozer, S.; Yang, L.L.; Novitch, B.; Martí, E.; et al. Distinct Sonic Hedgehog signaling dynamics specify floor plate and ventral neuronal progenitors in the vertebrate neural tube. Genes Dev. 2010, 24, 1186–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasai, N.; Kutejova, E.; Briscoe, J. Integration of signals along orthogonal axes of the vertebrate neural tube controls progenitor competence and increases cell diversity. PLoS Biol. 2014, 12, e1001907. [Google Scholar] [CrossRef] [PubMed]
- Diez del Corral, R.; Olivera-Martinez, I.; Goriely, A.; Gale, E.; Maden, M.; Storey, K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 2003, 40, 65–79. [Google Scholar] [CrossRef]
- Mathis, L.; Kulesa, P.M.; Fraser, S.E. FGF receptor signalling is required to maintain neural progenitors during hensen's node progression. Nat. Cell Biol. 2001, 3, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Delfino-Machin, M.; Lunn, J.S.; Breitkreuz, D.N.; Akai, J.; Storey, K.G. Specification and maintenance of the spinal cord stem zone. Development 2005, 132, 4273–4283. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Jeong, Y.; Misra, K.; Li, S.; Zelman, A.K.; Epstein, D.J.; Matise, M.P. Wnt signaling inhibitors regulate the transcriptional response to morphogenetic Shh-Gli signaling in the neural tube. Dev. Cell 2006, 11, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Gaiano, N.; Fishell, G. The role of notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci. 2002, 25, 471–490. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.H.; Yang, L.; Dessaud, E.; Chuang, K.; Moore, D.M.; Rohatgi, R.; Briscoe, J.; Novitch, B.G. Notch activity modulates the responsiveness of neural progenitors to Sonic hedgehog signaling. Dev. Cell 2015, 33, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Stasiulewicz, M.; Gray, S.D.; Mastromina, I.; Silva, J.C.; Bjorklund, M.; Seymour, P.A.; Booth, D.; Thompson, C.; Green, R.J.; Hall, E.A.; et al. A conserved role for Notch signaling in priming the cellular response to Shh through ciliary localisation of the key Shh transducer Smo. Development 2015, 142, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.K.; Vesque, C.; Lints, T.J.; Sampath, T.K.; Furley, A.; Dodd, J.; Placzek, M. Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell 1997, 90, 257–269. [Google Scholar] [CrossRef]
- Bourikas, D.; Pekarik, V.; Baeriswyl, T.; Grunditz, A.; Sadhu, R.; Nardo, M.; Stoeckli, E.T. Sonic hedgehog guides commissural axons along the longitudinal axis of the spinal cord. Nat. Neurosci. 2005, 8, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, S.A.; Tyurina, O.V.; Miller, E.; Bagas, A.; Karlstrom, R.O. Brother of cdo (umleitung) is cell-autonomously required for Hedgehog-mediated ventral CNS patterning in the zebrafish. Development 2011, 138, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Connor, R.M.; Allen, C.L.; Devine, C.A.; Claxton, C.; Key, B. BOC, brother of CDO, is a dorsoventral axon-guidance molecule in the embryonic vertebrate brain. J. Comp. Neurol. 2005, 485, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Charron, F.; Morin, S.; Shin, D.S.; Wong, K.; Fabre, P.J.; Tessier-Lavigne, M.; McConnell, S.K. Boc is a receptor for Sonic hedgehog in the guidance of commissural axons. Nature 2006, 444, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, M.F.; Damhofer, H.; Roelink, H. Hedgehog-stimulated chemotaxis is mediated by smoothened located outside the primary cilium. Sci. Signal. 2012, 5, ra60. [Google Scholar] [CrossRef] [PubMed]
- Kiecker, C.; Bates, T.; Bell, E. Molecular specification of germ layers in vertebrate embryos. Cell. Mol. Life Sci. 2016, 73, 923–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oosterveen, T.; Kurdija, S.; Alekseenko, Z.; Uhde, C.W.; Bergsland, M.; Sandberg, M.; Andersson, E.; Dias, J.M.; Muhr, J.; Ericson, J. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Dev. Cell 2012, 23, 1006–1019. [Google Scholar] [CrossRef] [PubMed]
- Oosterveen, T.; Kurdija, S.; Enstero, M.; Uhde, C.W.; Bergsland, M.; Sandberg, M.; Sandberg, R.; Muhr, J.; Ericson, J. SoxB1-driven transcriptional network underlies neural-specific interpretation of morphogen signals. Proc. Natl. Acad. Sci. USA 2013, 110, 7330–7335. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; Martinez Arias, A. Boundaries and fields in early embryos. Cell 1992, 68, 221–235. [Google Scholar] [CrossRef]
- Ingham, P.W. Localized hedgehog activity controls spatial limits of wingless transcription in the Drosophila embryo. Nature 1993, 366, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Cadigan, K.M.; Grossniklaus, U.; Gehring, W.J. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994, 8, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Buescher, M.; Svendsen, P.C.; Tio, M.; Miskolczi-McCallum, C.; Tear, G.; Brook, W.J.; Chia, W. Drosophila T box proteins break the symmetry of hedgehog-dependent activation of wingless. Curr. Biol. 2004, 14, 1694–1702. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; Placzek, M. Orchestrating ontogenesis: Variations on a theme by sonic hedgehog. Nat. Rev. Genet. 2006, 7, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Strigini, M.; Cohen, S.M. Formation of morphogen gradients in the Drosophila wing. Semin. Cell Dev. Biol. 1999, 10, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T. Genetics of morphogen gradients. Nat. Rev. Genet. 2001, 2, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Strigini, M.; Cohen, S.M. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 1997, 124, 4697–4705. [Google Scholar] [PubMed]
- Pappu, K.S.; Chen, R.; Middlebrooks, B.W.; Woo, C.; Heberlein, U.; Mardon, G. Mechanism of hedgehog signaling during Drosophila eye development. Development 2003, 130, 3053–3062. [Google Scholar] [CrossRef] [PubMed]
- Firth, L.C.; Baker, N.E. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 2005, 8, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Neumann, C.J. Hedgehogs as negative regulators of the cell cycle. Cell Cycle 2005, 4, 1139–1140. [Google Scholar] [CrossRef] [PubMed]
- Won, J.H.; Tsogtbaatar, O.; Son, W.; Singh, A.; Choi, K.W.; Cho, K.O. Cell type-specific responses to wingless, hedgehog and decapentaplegic are essential for patterning early eye-antenna disc in Drosophila. PLoS ONE 2015, 10, e0121999. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, D.; Kobayashi, M.; Matsumoto, K.; Ogura, T.; Nakafuku, M.; Shimamura, K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development 2002, 129, 83–93. [Google Scholar] [PubMed]
- Hashimoto-Torii, K.; Motoyama, J.; Hui, C.C.; Kuroiwa, A.; Nakafuku, M.; Shimamura, K. Differential activities of sonic hedgehog mediated by Gli transcription factors define distinct neuronal subtypes in the dorsal thalamus. Mech. Dev. 2003, 120, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Kiecker, C.; Lumsden, A. Hedgehog signaling from the ZLI regulates diencephalic regional identity. Nat. Neurosci. 2004, 7, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Scholpp, S.; Wolf, O.; Brand, M.; Lumsden, A. Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development 2006, 133, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Vue, T.Y.; Bluske, K.; Alishahi, A.; Yang, L.L.; Koyano-Nakagawa, N.; Novitch, B.; Nakagawa, Y. Sonic hedgehog signaling controls thalamic progenitor identity and nuclei specification in mice. J. Neurosci. 2009, 29, 4484–4497. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.; Garda, A.L.; Shimamura, K.; Martinez, S. Thalamic development induced by shh in the chick embryo. Dev. Biol. 2005, 284, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Szabo, N.E.; Zhao, T.; Zhou, X.; Alvarez-Bolado, G. The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J. Neurosci. 2009, 29, 2453–2466. [Google Scholar] [CrossRef] [PubMed]
- Robertshaw, E.; Matsumoto, K.; Lumsden, A.; Kiecker, C. Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc. Natl. Acad. Sci. USA 2013, 110, E3919–E3926. [Google Scholar] [CrossRef] [PubMed]
- Juraver-Geslin, H.A.; Gomez-Skarmeta, J.L.; Durand, B.C. The conserved barh-like homeobox-2 gene barhl2 acts downstream of orthodentricle-2 and together with iroquois-3 in establishment of the caudal forebrain signaling center induced by sonic hedgehog. Dev. Biol. 2014, 396, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Pierani, A.; Jessell, T.M.; Ericson, J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 2000, 101, 435–445. [Google Scholar] [CrossRef]
- Li, D.; Sakuma, R.; Vakili, N.A.; Mo, R.; Puviindran, V.; Deimling, S.; Zhang, X.; Hopyan, S.; Hui, C.C. Formation of proximal and anterior limb skeleton requires early function of Irx3 and Irx5 and is negatively regulated by Shh signaling. Dev. Cell 2014, 29, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Cavodeassi, F.; Diez del Corral, R.; Campuzano, S.; Dominguez, M. Compartments and organising boundaries in the Drosophila eye: The role of the homeodomain iroquois proteins. Development 1999, 126, 4933–4942. [Google Scholar] [PubMed]
- Cavodeassi, F.; Modolell, J.; Campuzano, S. The iroquois homeobox genes function as dorsal selectors in the Drosophila head. Development 2000, 127, 1921–1929. [Google Scholar] [PubMed]
- Calleja, M.; Herranz, H.; Estella, C.; Casal, J.; Lawrence, P.; Simpson, P.; Morata, G. Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development 2000, 127, 3971–3980. [Google Scholar] [PubMed]
- Calleja, M.; Renaud, O.; Usui, K.; Pistillo, D.; Morata, G.; Simpson, P. How to pattern an epithelium: Lessons from achaete-scute regulation on the notum of Drosophila. Gene 2002, 292, 1–12. [Google Scholar] [CrossRef]
- Ikmi, A.; Netter, S.; Coen, D. Prepatterning the Drosophila notum: The three genes of the iroquois complex play intrinsically distinct roles. Dev. Biol. 2008, 317, 634–648. [Google Scholar] [CrossRef] [PubMed]
- Mirzoyan, Z.; Pandur, P. The iroquois complex is required in the dorsal mesoderm to ensure normal heart development in Drosophila. PLoS ONE 2013, 8, e76498. [Google Scholar] [CrossRef]
- Matsumoto, K.; Nishihara, S.; Kamimura, M.; Shiraishi, T.; Otoguro, T.; Uehara, M.; Maeda, Y.; Ogura, K.; Lumsden, A.; Ogura, T. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat. Neurosci. 2004, 7, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Skarmeta, J.L.; Diez del Corral, R.; de la Calle-Mustienes, E.; Ferre-Marco, D.; Modolell, J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 1996, 85, 95–105. [Google Scholar] [CrossRef]
- Carrasco-Rando, M.; Tutor, A.S.; Prieto-Sanchez, S.; Gonzalez-Perez, E.; Barrios, N.; Letizia, A.; Martin, P.; Campuzano, S.; Ruiz-Gomez, M. Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLoS Genet. 2011, 7, e1002186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan, M.; Tabin, C.J. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science 1999, 283, 1736–1739. [Google Scholar] [CrossRef] [PubMed]
- DeLaurier, A.; Schweitzer, R.; Logan, M. Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev. Biol. 2006, 299, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Kiecker, C.; Niehrs, C. A morphogen gradient of Wnt/β-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 2001, 128, 4189–4201. [Google Scholar] [PubMed]
- Houart, C.; Caneparo, L.; Heisenberg, C.; Barth, K.; Take-Uchi, M.; Wilson, S. Establishment of the telencephalon during gastrulation by local antagonism of Wnt signaling. Neuron 2002, 35, 255–265. [Google Scholar] [CrossRef]
- Nordstrom, U.; Jessell, T.M.; Edlund, T. Progressive induction of caudal neural character by graded wnt signaling. Nat. Neurosci. 2002, 5, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.M.; Etheridge, A.; Bernard, A.; Robertson, C.P.; Roelink, H. Wnt signaling is required at distinct stages of development for the induction of the posterior forebrain. Development 2003, 130, 5579–5587. [Google Scholar] [CrossRef] [PubMed]
- Uygur, A.; Young, J.; Huycke, T.R.; Koska, M.; Briscoe, J.; Tabin, C.J. Scaling pattern to variations in size during development of the vertebrate neural tube. Dev. Cell 2016, 37, 127–135. [Google Scholar] [CrossRef] [PubMed]
| Tissue | Role | HH Ligand | Differential Competence? | References |
|---|---|---|---|---|
| Spinal cord | Ventral induction, later: growth, guidance of commissural axons, glia cell production | SHH | Floor plate vs. ventral interneuron induction (FGF-NKX1.2); motor neuron vs. oligodendrocyte induction (progenitor movement); patterning vs. growth (temporal adaptation, Notch signalling); axon guidance (receptor switch—HHIP, BOC, SMO localisation) | [45,48,93,94,96,97,100,101,103,104,105,106,107,111,112,113,116,118,119,121,122,123,124] |
| Cerebellum | Expansion | SHH | [66,67,68] | |
| Midbrain | Ventral induction (arcs), later: growth of tegmentum and tectum | SHH | Patterning vs. growth | [51,69] |
| Hypothalamus | Induction, patterning, expansion | SHH | Patterning vs. growth | [50,120] |
| Diencephalon | Growth, later: thalamus/prethalamus patterning | SHH | Growth/patterning; prethalamus vs. thalamus (PAX6 and IRX3) | [70,142,143,144,145,146,147,148] |
| Telencephalon | Subpallium induction, later: neocortex expansion | SHH | Patterning vs. growth (GLI downregulation by NKX2.1) | [98,102] |
| Early neural plate | Patterning | SHH | Anterior NKX2.1 vs. posterior FOXA2 induction (SIX3/IRX3) | [141] |
| CNS | Stem cell maintenance and activation in response to injury | SHH | [77,78,79,81,82] | |
| Limb bud | Anteroposterior patterning, growth | SHH | Forelimb vs. hindlimb (PITX1); patterning vs. growth | [53,99,151,161,162] |
| Somites | Sclerotome induction | SHH | [56,57,108] | |
| Muscle | Fibre induction | SHH | Slow-twitch vs. fast fibre (progenitor movement) | [109,110] |
| Pituitary gland | Induction | SHH | [58] | |
| Teeth | Induction | SHH | [63] | |
| Intestinal epithelium | Inhibition of pancreas induction, later: restriction of stem cell population, enterocyte differentiation | IHH, SHH | [59,75] | |
| Bladder epithelium | Regenerative proliferation | SHH | [83] | |
| Skin | Hair follicle development | SHH | [62] | |
| Lingual epithelium | Taste bud induction | SHH | [61] | |
| Germ line | Leydig cell differentiation, germ cell survival | DHH | [64] | |
| Skeleton | Cartilage differentiation | IHH | [65] | |
| Drosophila ectoderm | Segmental patterning | HH | Anterior: wg induction (slp vs. mid/h15) | [130,131,132] |
| Drosophila wing imaginal disc | Anteroposterior patterning | HH | Anterior: ptc/dpp induction | [136] |
| Drosophila eye imaginal disc | Photoreceptor differentiation | HH | [137,138,139] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiecker, C.; Graham, A.; Logan, M. Differential Cellular Responses to Hedgehog Signalling in Vertebrates—What is the Role of Competence? J. Dev. Biol. 2016, 4, 36. https://doi.org/10.3390/jdb4040036
Kiecker C, Graham A, Logan M. Differential Cellular Responses to Hedgehog Signalling in Vertebrates—What is the Role of Competence? Journal of Developmental Biology. 2016; 4(4):36. https://doi.org/10.3390/jdb4040036
Chicago/Turabian StyleKiecker, Clemens, Anthony Graham, and Malcolm Logan. 2016. "Differential Cellular Responses to Hedgehog Signalling in Vertebrates—What is the Role of Competence?" Journal of Developmental Biology 4, no. 4: 36. https://doi.org/10.3390/jdb4040036






