Abaxial Greening Phenotype in Hybrid Aspen
Abstract
:1. Introduction
1.1. Molecular Genetics of Leaf Variation
1.2. Objectives

2. Results and Discussion
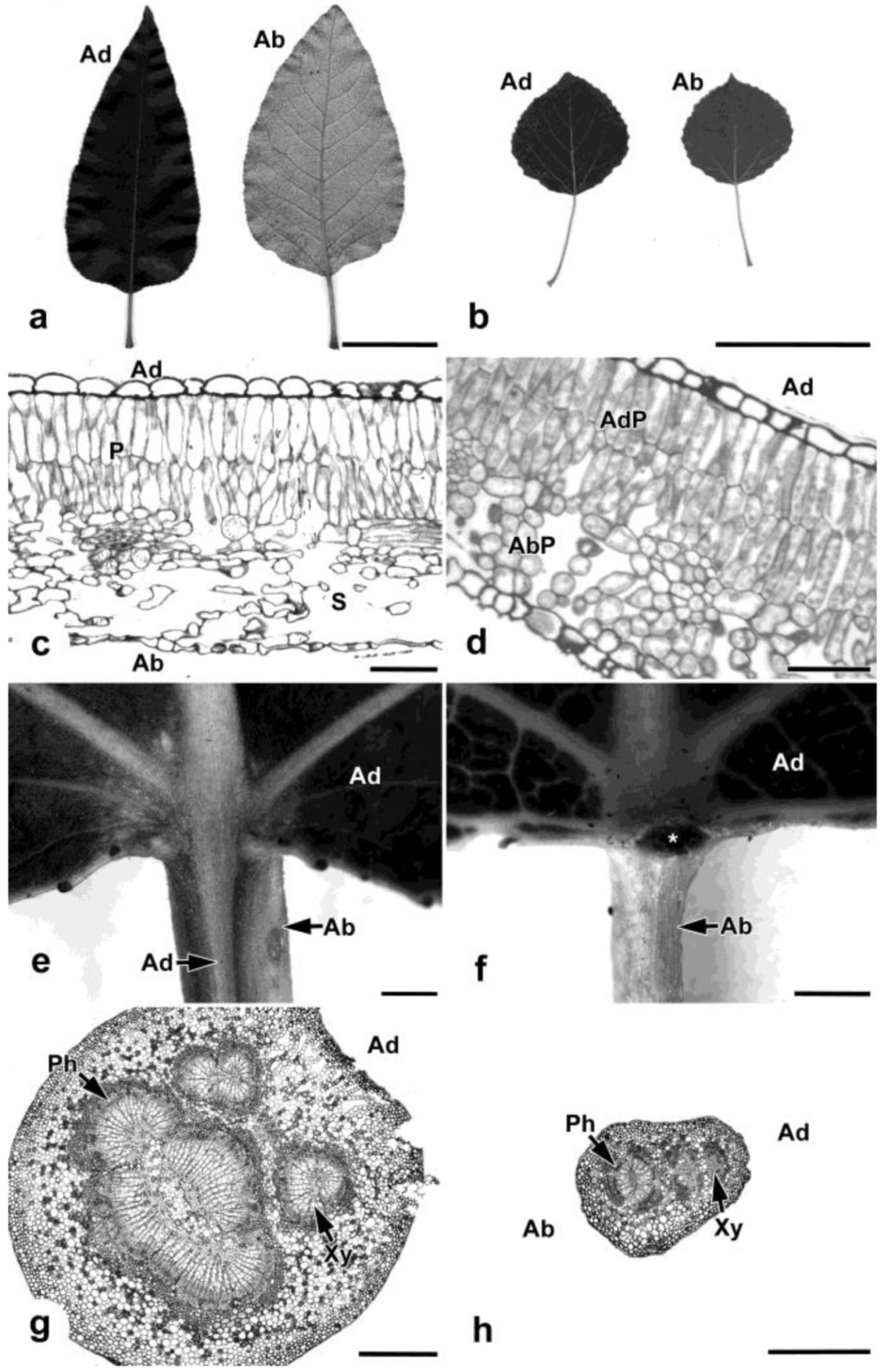
2.1. Leaf Analysis
2.2. Transcriptome Data Analysis
| Arabidopsis gene name | Arabidopsis accession number | Gene function in Arabidopsis | POPTR gene id (v2.2) | Poplar gene name |
|---|---|---|---|---|
| AE7 (AS1/AS2 enhancer 7) | AT1G68310 | Adaxial polarity formation | POPTR_0001s01820 | Pt-AE7.1 |
| POPTR_0003s09670 | Pt-AE7.2 | |||
| AGO1 (argonaute 1) | AT1G48410 | Adaxial/abaxial cell fate specification; vegetative phase change | POPTR_0012s03410 | Pt-AGO1.1 |
| POPTR_0015s05550 | Pt-AGO1.2 | |||
| AS1 (asymmetric leaves 1) | AT2G37630 | Adaxial axis specification | POPTR_0006s08610 | Pt-AS1.1 |
| POPTR_0004s10250 | Pt-AS1.2 | |||
| POPTR_0017s13950 | Pt-AS1.3 | |||
| AS2 (asymmetric leaves 2) | AT1G65620 | Adaxial axis specification | POPTR_0010s18460 | Pt-AS2.1 |
| POPTR_0008s07930 | Pt-AS2.2 | |||
| ATS (aberrant test shape) | AT5G42630 | Integument development; abaxial cell fate | POPTR_0002s13170 | Pt-ATS.1 |
| POPTR_0014s03650 | Pt-ATS.2 | |||
| YAB2 (YABBY 2) | AT1G08465 | Abaxial cell fate specification | POPTR_0001s22180 | Pt-YAB2.1 |
| POPTR_0127s00201 | Pt-YAB2.2 | |||
| POPTR_0016s06760 | Pt-YAB2.3 | |||
| YAB3 (YABBY 3) | AT4G00180 | Abaxial cell fate specification | POPTR_0003s11230 | Pt-YAB3.1 |
| POPTR_0001s00240 | Pt-YAB3.2 | |||
| ZPR3 (little zipper 3) | AT3G52770 | Adaxial cell fate specification | POPTR_0006s08320 | Pt-ZPR3.1 |
| POPTR_0010s24410 | Pt-ZPR3.2 |
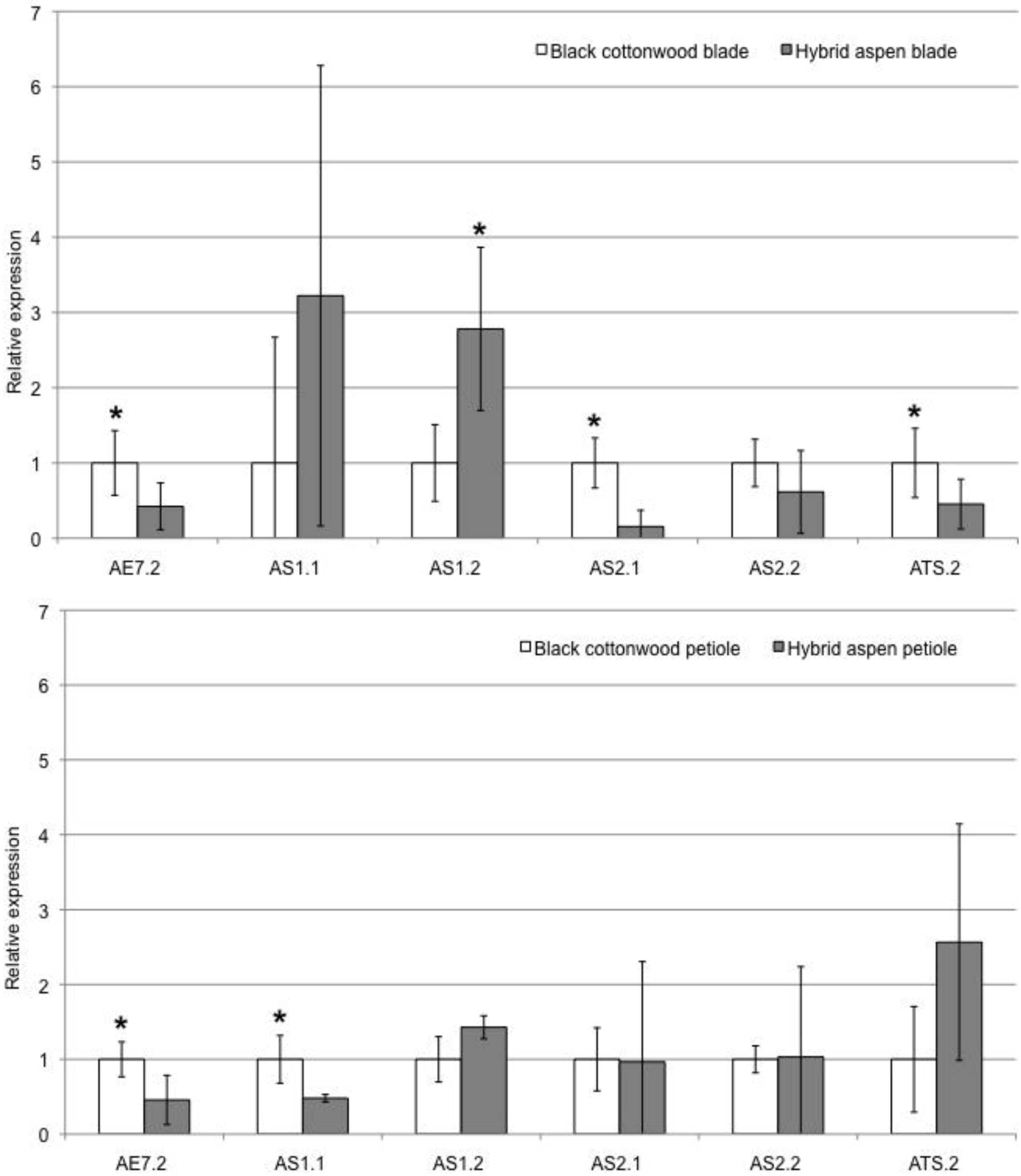
2.3. Leaf Blade and Petiole qRT-PCR Gene Expression Patterns
| POPTR gene id (v2.2) | Gene name | Hybrid or European aspen abundance in relation to black cottonwood | |||
|---|---|---|---|---|---|
| mRNA-seq | RT-PCR | qRT-PCR | qRT-PCR p-value | ||
| POPTR_0003s09670 | Pt-AE7.2 | - | - | - | 0.020 |
| POPTR_0015s05550 | Pt-AGO1.2 | NS | + | - * | 0.008 |
| POPTR_0004s10250 | Pt-AS1.2 | NS | NA | + * | 0.009 |
| POPTR_0010s18460 | Pt-AS2.1 | - | - | - | 0.028 |
| POPTR_0014s03650 | Pt-ATS.2 | + | + | - * | 0.030 |
| POPTR_0001s22180 | Pt-YAB2.1 | - | NA | - * | 0.010 |
| POPTR_0001s00240 | Pt-YAB3.2 | + | + | + | 0.00008 |
| POPTR_0006s08320 | Pt-ZPR3.1 | - | - | - | 0.020 |
| POPTR_0010s24410 | Pt-ZPR3.2 | + | + | - | 0.005 |
2.4. Abaxial Determinants in Aspen
2.5. ARGONAUTE1 in Aspen
2.6. ASYMMETRIC LEAVES1 in Aspen
3. Experimental Section
3.1. Leaf Analysis
3.2. Gene Selection
3.3. mRNA-seq Gene Expression Data Analysis
3.4. Reverse Transcriptase PCR
3.5. Quantitative RT-PCR
4. Conclusions
Acknowledgments
References
- Eckenwalder, J.E. Systematics and evolution of Populus. In Biology of Populus and Its Implications for Management and Conservation; Stettler, R.F., Bradshaw, H.D., Jr., Heilman, P.E., Hinckley, T.M., Eds.; NRC Research Press, National Research Council of Canada: Ottawa, Canada, 1996; pp. 7–32. [Google Scholar]
- Van Volkenburgh, E.; Taylor, G. Leaf growth physiology. In Biology of Populus and Its Implications for Management and Conservation; Stettler, R.F., Bradshaw, H.D., Jr., Heilman, P.E., Hinckley, T.M., Eds.; NRC Research Press, National Research Council of Canada: Ottawa, Canada, 1996; pp. 283–299. [Google Scholar]
- Cronk, Q.C.B. Plant eco-devo: The potential of poplar as a model organism. New Phytol. 2005, 166, 39–48. [Google Scholar] [CrossRef]
- Wu, R.; Bradshaw, H.D., Jr.; Stettler, R.F. Molecular genetics of growth and development in Populus (Salicaceae). V. Mapping quantitative trait loci affecting leaf variation. Am. J. Bot. 1997, 84, 143–153. [Google Scholar] [CrossRef]
- Roden, J.S.; Pearcy, R.W. Effect of leaf flutter on the light environment in poplars. Oecologia 1993, 93, 201–207. [Google Scholar] [CrossRef]
- Roden, J.S.; Pearcy, R.W. The effect of leaf flutter on the flux of CO2 in poplar leaves. Funct. Ecol. 1993, 7, 669–675. [Google Scholar] [CrossRef]
- Roden, J.S.; Pearcy, R.W. Photosynthetic gas exchange response of poplars to steady-state and dynamic light environments. Oecologia 1993, 93, 208–214. [Google Scholar] [CrossRef]
- Wang, J.W.; Park, M.Y.; Wang, L.J.; Koo, Y.; Chen, X.Y.; Weigel, D.; Poethig, R.S. MiRNA control of vegetative phase change in trees. PLoS Genet. 2011, 7, e1002012. [Google Scholar]
- Waites, R.; Hudson, A. phantastica: A gene required for dorsoventrality of leaves in Antiffhinum majus. Development 1995, 121, 2143–2154. [Google Scholar]
- Kim, M.; McCormick, S.; Timmermans, M.; Sinha, N. The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 2003, 424, 438–443. [Google Scholar] [CrossRef]
- McHale, N.A.; Koning, R.E. PHANTASTICA regulates development of the adaxial mesophyll in Nicotiana leaves. Plant Cell 2004, 16, 1251–1262. [Google Scholar] [CrossRef]
- Zoulias, N.; Koenig, D.; Hamidi, A.; McCormick, S.; Kim, M. A role for PHANTASTICA in medio-lateral regulation of adaxial domain development in tomato and tobacco leaves. Ann. Bot. 2012, 109, 407–418. [Google Scholar] [CrossRef]
- Chua, Y.L.; Channeliere, S.; Mott, E.; Gray, J.C. The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev. 2005, 19, 2245–2254. [Google Scholar] [CrossRef]
- Kidner, C.A.; Timmermans, M.C. Signaling sides: Adaxial-abaxial patterning in leaves. Curr. Top. Dev. Biol. 2010, 91, 141–168. [Google Scholar] [CrossRef]
- Townsley, B.T.; Sinha, N.R. A new development: Evolving concepts in leaf ontogeny. Annu. Rev. Plant Biol. 2012, 63, 535–562. [Google Scholar] [CrossRef]
- Yuan, Z.; Luo, D.; Li, G.; Yao, X.; Wang, H.; Zeng, M.; Huang, H.; Cui, X. Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J. 2010, 64, 331–342. [Google Scholar] [CrossRef]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Scholkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef]
- McAbee, J.M.; Hill, T.A.; Skinner, D.J.; Izhaki, A.; Hauser, B.A.; Meister, R.J.; Reddy, G.V.; Meyerowitz, E.K.; Bowman, J.L.; Gasser, C.S. ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 2006, 46, 522–531. [Google Scholar] [CrossRef]
- Kelley, D.R.; Skinner, D.J.; Gasser, C.S. Roles of polarity determinants in ovule development. Plant J. 2009, 57, 1054–1064. [Google Scholar] [CrossRef]
- Gao, P.; Li, X.; Cui, D.; Wu, L.; Parkin, I.; Gruber, M.Y. A new dominant Arabidopsis transparent testa mutant, sk21-D, and modulation of seed flavonoid biosynthesis by KAN4. Plant Biotechnol. J. 2010, 8, 979–993. [Google Scholar] [CrossRef]
- Nakagawa, A.; Takahashi, H.; Kojima, S.; Sato, N.; Ohga, K.; Cha, B.Y.; Woo, J.T.; Nagai, K.; Horiguchi, G.; Tsukaya, H.; et al. Berberine enhances defects in the establishment of leaf polarity in asymmetric leaves1 and asymmetric leaves2 of Arabidopsis thaliana. Plant Mol. Biol. 2012, 79, 569–581. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, D.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 128, 4117–4128. [Google Scholar]
- Bowman, J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000, 3, 17–22. [Google Scholar] [CrossRef]
- Nowak, J.S. Effects of Adaxial-Abaxial Signalling on Leaf Polarity. Ph.D. Dissertation, University of British Columbia, Vancouver, Canada, 2012. [Google Scholar]
- Sawa, S.; Watanabe, K.; Goto, K.; Liu, Y.G.; Shibata, D.; Kanaya, E.; Morita, E.H.; Okada, K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG related domains. Genes Dev. 1999, 13, 1079–1088. [Google Scholar] [CrossRef]
- Eshed, Y.; Baum, S.F.; Perea, J.V.; Bowman, J.L. Establishment of polarity in lateral organs of plants. Curr.Biol. 2001, 11, 1251–1260. [Google Scholar] [CrossRef]
- Szakonyi, D.; Moschopoulos, A.; Byrne, M.E. Perspectives on leaf dorsoventral polarity. J. Plant Res. 2010, 123, 281–290. [Google Scholar] [CrossRef]
- Kidner, C.A.; Martienssen, R.A. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 2004, 428, 81–84. [Google Scholar] [CrossRef]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONATUE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef]
- Lynn, K.; Fernandez, A.; Aida, M.; Sedbrook, J.; Tasaka, M.; Masson, P.; Barton, M.K. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 1999, 126, 469–481. [Google Scholar]
- Bohmert, K.; Camus, I.; Bellini, C.; Bouchez, D.; Caboche, M.; Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998, 17, 170–180. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Pi, L.; Liu, Q.; Ling, Q.; Wang, H.; Poethig, R.S.; Huang, H. Genetic interaction between the AS1-AS2 and RDR6-SGS3-AGO7 pathways for leaf morphogenesis. Plant Cell Physiol. 2006, 47, 853–863. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Z.; Lu, F.; Dong, A.; Huang, H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006, 47, 841–850. [Google Scholar] [CrossRef]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar]
- Byrne, M.E.; Barley, R.; Curtis, M.; Arroyo, J.M.; Dunham, M.; Hudson, A.; Martienssen, R.A. ASYMMETRIC LEAVES1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef]
- Lin, W.C.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef]
- Kojima, S.; Iwasaki, M.; Takahashi, H.; Imai, T.; Matsumura, Y.; Fleury, D.; Lijsebettens, M.V.; Machida, Y.; Machida, C. ASYMMERTIC LEAVES2 and elongator, a histone acetyltransferase complex, mediate the extablishment ofpolarity in leaves of Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1259–1273. [Google Scholar] [CrossRef]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES 2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef]
- Byrne, M.E. Networks in leaf development. Curr. Opin. Plant Biol. 2005, 8, 59–66. [Google Scholar] [CrossRef]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef]
- Li, H.; Xu, L.; Wang, H.; Yuan, Z.; Cao, X.; Yang, Z.; Zhang, D.; Xu, Y.; Huang, H. The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 2005, 17, 2157–2171. [Google Scholar] [CrossRef]
- Fu, Y.; Xu, L.; Xu, B.; Yang, L.; Ling, Q.; Wang, H.; Huang, H. Genetic interaction between leaf polarity-controlling genes ASYMMETRIC LEAVES1 and 2 in Arabidopsis leaf patterning. Plant Cell Physiol. 2007, 48, 724–735. [Google Scholar] [CrossRef]
- Nowak, J.; Dengler, N.G.; Posluszny, U. The role of abscission during leaflet separation in Chamaedorea elegans (Arecaceae). Int. J. Plant Sci. 2007, 168, 533–545. [Google Scholar] [CrossRef]
- Arabidopsis Information Resource (TAIR). Available online: http://www.arabidopsis.org/ (accessed on 30 September 2011).
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar]
- Geraldes, A.; Pang, J.; Thiessen, N.; Cezard, T.; Moore, R.; Zhao, Y.; Tam, A.; Wang, S.; Friedmann, M.; Birol, I.; et al. SNP discovery in black cottonwood (Populus trichocarpa) by population transcriptome resequencing. Mol. Ecol. Resour. 2011, 11, 81–92. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Rozen, S.; Skaletsky, H.J. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology; Krawetz, S., Misener, S., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 365–386. [Google Scholar]
- Ralph, S.; Oddy, C.; Cooper, D.; Yueh, H.; Jancsik, S.; Kolosova, N.; Philippe, R.N.; Aeschliman, D.; White, R.; Huber, D.; et al. Genomics of hybrid poplar (Populus trichocarpa x deltoides) interacting with forest tent caterpillars (Malacosoma disstria): Normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defenses in poplar. Mol. Ecol. 2006, 15, 1275–1297. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar]
- Pfaffl, M.W. Relative quantification. In Real-Time PCR; Dorak, M.T., Ed.; Taylor & Francis Group: New York, NY, USA, 2006; pp. 63–82. [Google Scholar]
Supplementary Materials
| Arabidopsis gene name | Arabidopsis accession number | Gene function in Arabidopsis | POPTR gene id (v2.2) | Poplar gene name |
|---|---|---|---|---|
| AE3 (AS1/AS2 enhancer 3) | AT5G05780 | Adaxial leaf identity specification | POPTR_0008s06540 | Pt-AE3.1 |
| POPTR_0008s06550 | Pt-AE3.2 | |||
| AE7 (AS1/2 enhancer 7) | AT1G68310 | Adaxial polarity formation | POPTR_0001s01820 | Pt-AE7.1 |
| POPTR_0003s09670 | Pt-AE7.2 | |||
| AFO (abnormal flower organ) | AT2G45190 | Abaxial cell fate specification | POPTR_0014s06210 | Pt-AFO.1 |
| POPTR_0002s14600 | Pt-AFO.2 | |||
| AGO1 (argonaute 1) | AT1G48410 | Adaxial/abaxial cell fate specification; vegetative phase change | POPTR_0012s03410 | Pt-AGO1.1 |
| POPTR_0015s05550 | Pt-AGO1.2 | |||
| AGO7 (argonaute 7) | AT1G69440 | Regulation of vegetative phase change | POPTR_0009s00660 | Pt-AGO7.1 |
| POPTR_0010s17100 | Pt-AGO7.4 | |||
| AGO10 (argonaute 10) | AT5G43810 | Primary SAM specification; miRNA binding | POPTR_0008s15860 | Pt-AGO10.1 |
| POPTR_0010s09150 | Pt-AGO10.2 | |||
| AN3 (angustifolia 3) | AT5G28640 | Leaf development | POPTR_0013s04090 | Pt-AN3.1 |
| POPTR_0019s02320 | Pt-AN3.2 | |||
| ARF4 (auxin response factor 4) | AT5G60450 | Abaxial cell fate specification | POPTR_0009s01700 | Pt-ARF4.1 |
| AS1 (asymmetric leaves 1) | AT2G37630 | Adaxial axis specification | POPTR_0006s08610 | Pt-AS1.1 |
| POPTR_0004s10250 | Pt-AS1.2 | |||
| POPTR_0017s13950 | Pt-AS1.3 | |||
| AS2 (asymmetric leaves 2) | AT1G65620 | Adaxial axis specification | POPTR_0010s18460 | Pt-AS2.1 |
| POPTR_0008s07930 | Pt-AS2.2 | |||
| CNA (corona) | AT1G52150 | Adaxial identity determination; vascular histogenesis | POPTR_0003s04860 | Pt-ATHB.11 |
| POPTR_0001s18930 | Pt-ATHB.12 | |||
| ATS (aberrant test shape) | AT5G42630 | Integument development; abaxial cell fate | POPTR_0002s13170 | Pt-ATS.1 |
| POPTR_0014s03650 | Pt-ATS.2 | |||
| CRC (crabs claw) | AT1G69180 | Abaxial axis specification; floral meristem determinacy | POPTR_0008s09740 | Pt-CRC.1 |
| POPTR_0010s16410 | Pt-CRC.2 | |||
| DCL4 (dicer-like 4) | AT5G20320 | Vegetative phase change | POPTR_0006s20310 | Pt-DCL4.1 |
| DUF59 (domain of unknown function 59) | AT3G50845 | Adaxial polarity formation (AE7-like) | POPTR_0005s12480 | Pt-DUF59.1 |
| ETT (ettin) | AT2G33860 | Abaxial cell fate specification | POPTR_0004s04970 | Pt-ETT.1 |
| POPTR_0011s05830 | Pt-ETT.2 | |||
| ATHB8 (Arabidopsis thaliana homeobox 8) | AT4G32880 | Xylem development | POPTR_0018s08110 | Pt-HB1.5 |
| POPTR_0006s25390 | Pt-HB1.6 | |||
| REV (revoluta) | AT5G60690 | Adaxial axis specification; vascular pattern formation | POPTR_0004s22090 | Pt-HB1.7 |
| POPTR_0009s01990 | Pt-HB1.8 | |||
| HYL (hyponastic leaves 1) | AT1G09700 | Vegetative phase control (Li et al. 2012) | POPTR_0005s19650 | Pt-HYL1.1 |
| POPTR_0002s11200 | Pt-HYL1.2 | |||
| INO (inner-no-outer) | AT1G23420 | Abaxial cell fate specification; ovule development | POPTR_0008s19330 | Pt-INO.1 |
| POPTR_0010s05220 | Pt-INO.2 | |||
| KAN (KANADI 1) | AT5G16560 | Abaxial identity specification | POPTR_0017s02220 | Pt-KAN.1 |
| POPTR_0004s08070 | Pt-KAN.2 | |||
| POPTR_0015s05340 | Pt-KAN.3 | |||
| POPTR_0012s03900 | Pt-KAN.4 | |||
| KAN2, KAN3 (KANADI 2, KANADI3) | AT1G32240 AT4G17695 | Abaxial cell fate specification; ovule development | POPTR_0003s09490 | Pt-KAN2/3.1 |
| POPTR_0001s02010 | Pt-KAN2/3.2 | |||
| PHB (phabulosa)PHV (phavoluta) | AT2G34710 AT1G30490 | Adaxial cell fate specification | POPTR_0011s10070 | Pt-PHB.1 |
| POPTR_0001s38120 | Pt-PHB.2 | |||
| PGY1 (piggyback 1) | AT2G27530 | Adaxial pattern specification; AS1 enhancer | POPTR_0007s11880 | Pt-PGY1.1 |
| PGY2 (piggyback 2) | AT1G33140 | Adaxial pattern specification; AS1 enhancer | POPTR_0001s45810 | Pt-PGY2.1 |
| POPTR_0011s15170 | Pt-PGY2.2 | |||
| PGY3 (piggyback 3) | AT3G25520 | Adaxial pattern specification | POPTR_0013s13220 | Pt-PGY3.1 |
| RDR6 (RNA-dependent RNA polymerase 6) | AT3G49500 | Leaf development | POPTR_0006s26980 | Pt-RDR6.1 |
| POPTR_0018s01670 | Pt-RDR6.2 | |||
| SE (serrate) | AT2G27100 | Adaxial/abaxial pattern regulation | POPTR_0004s20730 | Pt-SE.1 |
| POPTR_0009s16020 | Pt-SE.2 | |||
| SGS3 (suppressor of gene silencing 3) | AT5G23570 | Vegetative phase change | POPTR_0019s00300 | Pt-SGS3.1 |
| POPTR_0001s07410 | Pt-SGS3.2 | |||
| POPTR_0001s07420 | Pt-SGS3.3 | |||
| POPTR_0003s18660 | Pt-SGS3.4 | |||
| POPTR_0003s18670 | Pt-SGS3.5 | |||
| POPTR_0003s18680 | Pt-SGS3.6 | |||
| POPTR_0003s18690 | Pt-SGS3.7 | |||
| POPTR_0003s01530 | Pt-SGS3.8 | |||
| SPL4 (squamosa promoter binding-like 4) | AT1G53160 | Vegetative phase change | POPTR_0001s40870 | Pt-SPL4.1 |
| POPTR_0011s11770 | Pt-SPL4.2 | |||
| SPL4, SPL3 (squamosa promoter binding-like 3) | AT2G33810 (SPL3) | Vegetative phase change regulation | POPTR_0004s04630 | Pt-SPL43.1 |
| POPTR_0011s05480 | Pt-SPL43.2 | |||
| SPL9 (squamosa promoter binding-like 9) | AT2G42200 | Vegetative to reproductive phase change transition | POPTR_0016s04890 | Pt-SPL9.1 |
| YAB2 (YABBY 2) | AT1G08465 | Abaxial cell fate specification | POPTR_0001s22180 | Pt-YAB2.1 |
| POPTR_0127s00201 | Pt-YAB2.2 | |||
| POPTR_0016s06760 | Pt-YAB2.3 | |||
| YAB3 (YABBY 3) | AT4G00180 | Abaxial cell fate specification | POPTR_0003s11230 | Pt-YAB3.1 |
| POPTR_0001s00240 | Pt-YAB3.2 | |||
| YAB5 (YABBY 5) | AT2G26580 | Abaxial cell fate specification | POPTR_0006s06700 | Pt-YAB5.1 |
| Abaxial cell fate specification | POPTR_0018s12990 | Pt-YAB5.2 | ||
| YUC (yucca) | AT4G32540 | Auxin biosynthesis; regulation of leaf development | POPTR_0006s26430 | Pt-YUC.2 |
| POPTR_0018s01210 | Pt-YUC.1 | |||
| YUC2 (yucca 2) | AT4G13260 | Auxin biosynthesis | POPTR_0006s26000 | Pt-YUC2.1 |
| POPTR_0018s00840 | Pt-YUC2.2 | |||
| ZPR1 (little zipper 1) | AT2G45450 | Adaxial cell fate specification; interacts with REV | POPTR_0003s11710 | Pt-ZPR1.1 |
| POPTR_0001s08220 | Pt-ZPR1.2 | |||
| ZPR2 (little zipper 2) | AT3G60890 | Adaxial cell fate specification | POPTR_0002s15060 | Pt-ZPR2.1 |
| POPTR_0014s06690 | Pt-ZPR2.2 | |||
| ZPR3 (little zipper 3) | AT3G52770 | Adaxial cell fate specification | POPTR_0006s08320 | Pt-ZPR3.1 |
| POPTR_0010s24410 | Pt-ZPR3.2 |
| Gene id | Gene name | Ptr mean RPKM | Ptmx mean RPKM | p-value |
|---|---|---|---|---|
| POPTR_0008s06540 | Pt-AE3.1 | 57.44 ± 8.16 | 40.04 ± 6.03 | 0.0221 |
| POPTR_0008s06550 | Pt-AE3.2 | 130.53 ± 7.99 | 95.61 ± 12.10 | 0.0077 |
| POPTR_0001s01820 | Pt-AE7.1 | 14.13 ± 4.50 | 11.69 ± 0.74 | 0.3228 |
| POPTR_0003s09670 | Pt-AE7.2 * | 26.16 ± 6.35 | 16.34 ± 1.34 | 0.0268 |
| POPTR_0014s06210 | Pt-AFO.1 | 98.67 ± 16.85 | 52.38 ± 25.72 | 0.0437 |
| POPTR_0002s14600 | Pt-AFO.2 | 115.90 ± 22.05 | 58.60 ± 24.51 | 0.0244 |
| POPTR_0012s03410 | Pt-AGO1.1 | 68.79 ± 21.07 | 80.39 ± 13.30 | 0.4085 |
| POPTR_0015s05550 | Pt-AGO1.2* | 32.07 ± 11.43 | 30.67 ± 6.36 | 0.8422 |
| POPTR_0009s00660 | Pt-AGO7.1 | 7.09 ± 2.27 | 7.05 ± 0.88 | 0.9738 |
| POPTR_0010s17100 | Pt-AGO7.4 * | 29.99 ± 5.89 | 24.16 ± 14.50 | 0.5471 |
| POPTR_0008s15860 | Pt-AGO10.1 * | 12.84 ± 5.27 | 21.34 ± 6.48 | 0.1241 |
| POPTR_0010s09150 | Pt-AGO10.2 | 30.49 ± 10.63 | 17.88 ± 8.20 | 0.1343 |
| POPTR_0013s04090 | Pt-AN3.1 | 44.23 ± 11.87 | 33.86 ± 14.80 | 0.3674 |
| POPTR_0019s02320 | Pt-AN3.2 | 94.75 ± 41.53 | 76.91 ± 43.04 | 0.6057 |
| POPTR_0009s01700 | Pt-ARF4.1 | 25.40 ± 9.95 | 24.31 ± 4.08 | 0.8477 |
| POPTR_0006s08610 | Pt-AS1.1 * | 52.68 ± 7.63 | 82.05 ± 41.07 | 0.2857 |
| POPTR_0004s10250 | Pt-AS1.2 * | 25.90 ± 13.53 | 7.36 ± 5.93 | 0.0544 |
| POPTR_0017s13950 | Pt-AS1.3 | 51.14 ± 9.22 | 40.39 ± 11.10 | 0.2334 |
| POPTR_0010s18460 | Pt-AS2.1 * | 23.81 ± 5.22 | 9.81 ± 6.46 | 0.0282 |
| POPTR_0008s07930 | Pt-AS2.2 * | 4.85 ± 0.24 | 1.07 ± 0.75 | 0.0004 |
| POPTR_0003s04860 | Pt-ATHB.11 | 13.90 ± 4.42 | 16.57 ± 6.17 | 0.5555 |
| POPTR_0001s18930 | Pt-ATHB.12 | 14.90 ± 4.21 | 36.77 ± 12.95 | 0.0399 |
| POPTR_0002s13170 | Pt-ATS.1 | 5.59 ± 0.60 | 4.54 ± 2.29 | 0.4815 |
| POPTR_0014s03650 | Pt-ATS.2 * | 13.96 ± 3.14 | 35.83 ± 7.50 | 0.0055 |
| POPTR_0008s09740 | Pt-CRC.1 | 0.00 | 0.02 ± 0.03 | 0.2751 |
| POPTR_0010s16410 | Pt-CRC.2 | 0.00 | 0.00 | n/a |
| POPTR_0006s20310 | Pt-DCL4.1 | 2.54 ± 1.04 | 4.97 ± 1.47 | 0.0592 |
| POPTR_0005s12480 | Pt-DUF59.1 | 23.32 ± 9.89 | 21.87 ± 3.62 | 0.7928 |
| POPTR_0004s04970 | Pt-ETT.1 | 17.98 ± 5.65 | 20.09 ± 2.29 | 0.5199 |
| POPTR_0011s05830 | Pt-ETT.2 | 6.33 ± 1.56 | 8.77 ± 2.76 | 0.2322 |
| POPTR_0018s08110 | Pt-HB1.5 (ATHB8) | 6.76 ± 2.39 | 19.63 ± 6.46 | 0.0234 |
| POPTR_0006s25390 | Pt-HB1.6 (ATHB8) | 32.29 ± 7.56 | 23.22 ± 3.89 | 0.0897 |
| POPTR_0004s22090 | Pt-HB1.7 (REV) | 6.04 ± 2.20 | 10.26 ± 3.55 | 0.1330 |
| POPTR_0009s01990 | Pt-HB1.8 (REV) | 25.92 ± 8.83 | 9.02 ± 1.62 | 0.0118 |
| POPTR_0005s19650 | Pt-HYL1.1 | 16.41 ± 5.81 | 19.42 ± 2.47 | 0.3851 |
| POPTR_0002s11200 | Pt-HYL1.2 | 5.71 ± 1.29 | 8.58 ± 0.69 | 0.0121 |
| POPTR_0008s19330 | Pt-INO.1 | 0.01 ± 0.01 | 0.00 | 0.2856 |
| POPTR_0010s05220 | Pt-INO.2 | 0.01 ± 0.02 | 0.00 | 0.2856 |
| POPTR_0017s02220 | Pt-KAN.1 | 4.80 ± 0.39 | 7.01 ± 5.89 | 0.5545 |
| POPTR_0004s08070 | Pt-KAN.2 | 15.22 ± 1.36 | 9.80 ± 0.67 | 0.0009 |
| POPTR_0015s05340 | Pt-KAN.3 | 2.61 ± 1.64 | 6.26 ± 1.94 | 0.0471 |
| POPTR_0012s03900 | Pt-KAN.4 | 3.27 ± 2.60 | 2.48 ± 0.85 | 0.5828 |
| POPTR_0003s09490 | Pt-KAN2/3.1 | 8.34 ± 0.93 | 1.08 ± 0.29 | 2.281 × 10−05 |
| POPTR_0001s02010 | Pt-KAN2/3.2 | 5.48 ± 1.11 | 7.81 ± 2.46 | 0.1925 |
| POPTR_0011s10070 | Pt-PHB.1 | 27.16 ± 5.37 | 45.01 ± 8.31 | 0.0238 |
| POPTR_0001s38120 | Pt-PHB.2 | 25.69 ± 9.86 | 8.84 ± 1.02 | 0.0171 |
| POPTR_0007s11880 | Pt-PGY1.1 | 197.62 ± 39.30 | 99.77 ± 34.79 | 0.0174 |
| POPTR_0001s45810 | Pt-PGY2.1 | 61.44 ± 25.32 | 41.11 ± 19.57 | 0.2812 |
| POPTR_0011s15170 | Pt-PGY2.2 | 129.11 ± 57.05 | 63.09 ± 17.23 | 0.0746 |
| POPTR_0013s13220 | Pt-PGY3.1 * | 406.38 ± 60.21 | 428.42 ± 107.56 | 0.7654 |
| POPTR_0006s26980 | Pt-RDR6.1 * | 5.83 ± 2.12 | 15.81 ± 1.06 | 0.0004 |
| POPTR_0018s01670 | Pt-RDR6.2 * | 1.13 ± 0.39 | 8.05 ± 2.57 | 0.0063 |
| POPTR_0004s20730 | Pt-SE.1 | 19.68 ± 2.93 | 24.42 ± 3.09 | 0.0956 |
| POPTR_0009s16020 | Pt-SE.2 | 25.06 ± 2.54 | 26.09 ± 5.93 | 0.7938 |
| POPTR_0019s00300 | Pt-SGS3.1 | 49.70 ± 3.60 | 44.12 ± 10.69 | 0.4333 |
| POPTR_0001s07410 | Pt-SGS3.2 | 20.06 ± 9.14 | 15.20 ± 15.20 | 0.3650 |
| POPTR_0001s07420 | Pt-SGS3.3 | 0.27 ± 0.22 | 0.39 ± 0.07 | 0.3351 |
| POPTR_0003s18660 | Pt-SGS3.4 | 2.06 ± 0.65 | 1.84 ± 0.54 | 0.6411 |
| POPTR_0003s18670 | Pt-SGS3.5 | 0.30 ± 0.07 | 0.10 ± 0.04 | 0.0061 |
| POPTR_0003s18680 | Pt-SGS3.6 | 3.56 ± 0.07 | 2.99 ± 0.89 | 0.4061 |
| POPTR_0003s18690 | Pt-SGS3.7 | 0.41 ± 0.14 | 0.15 ± 0.06 | 0.0188 |
| POPTR_0003s01530 | Pt-SGS3.8 | 2.22 ± 1.37 | 9.80 ± 6.01 | 0.0898 |
| POPTR_0001s40870 | Pt-SPL4.1 * | 28.12 ± 3.72 | 60.65 ± 8.68 | 0.0019 |
| POPTR_0011s11770 | Pt-SPL4.2 | 9.61 ± 0.10 | 11.10 ± 7.10 | 0.7370 |
| POPTR_0004s04630 | Pt-SPL43.1 | 2.70 ± 0.96 | 9.59 ± 9.15 | 0.2605 |
| POPTR_0011s05480 | Pt-SPL43.2 * | 51.31 ± 15.58 | 65.37 ± 24.85 | 0.4335 |
| POPTR_0016s04890 | Pt-SPL9.1 | 4.74 ± 1.95 | 9.24 ± 4.78 | 0.1914 |
| POPTR_0001s22180 | Pt-YAB2.1 | 102.90 ± 13.76 | 1.97 ± 1.53 | 2.352 × 10−05 |
| POPTR_0127s00201 | Pt-YAB2.2 | 16.82 ± 5.87 | 58.12 ± 30.25 | 0.0717 |
| POPTR_0016s06760 | Pt-YAB2.3 | 75.42 ± 16.33 | 64.62 ± 29.43 | 0.5962 |
| POPTR_0003s11230 | Pt-YAB3.1 | 61.42 ± 10.31 | 114.00 ± 51.14 | 0.1470 |
| POPTR_0001s00240 | Pt-YAB3.2 * | 14.74 ± 2.50 | 66.94 ± 10.40 | 0.0004 |
| POPTR_0006s06700 | Pt-YAB5.1 | 47.58 ± 19.28 | 60.05 ± 2.61 | 0.2434 |
| POPTR_0018s12990 | Pt-YAB5.2 | 220.13 ± 18.32 | 0.01 ± 0.01 | 1.959 × 10−06 |
| POPTR_0006s26430 | Pt-YUC.2 | 1.35 ± 0.63 | 1.08 ± 0.77 | 0.6348 |
| POPTR_0018s01210 | Pt-YUC.1 | 3.45 ± 1.59 | 1.27 ± 0.88 | 0.0652 |
| POPTR_0006s26000 | Pt-YUC2.1 | 2.91 ± 0.39 | 4.94 ± 1.81 | 0.1211 |
| POPTR_0018s00840 | Pt-YUC2.2 | 1.01 ± 0.67 | 0.76 ± 0.45 | 0.5844 |
| POPTR_0003s11710 | Pt-ZPR1.1 | 2.81 ± 1.35 | 2.76 ± 0.68 | 0.9491 |
| POPTR_0001s08220 | Pt-ZPR1.2 | 3.20 ± 1.47 | 1.47 ± 0.65 | 0.0845 |
| POPTR_0002s15060 | Pt-ZPR2.1 | 4.29 ± 3.00 | 3.35 ± 1.11 | 0.5780 |
| POPTR_0014s06690 | Pt-ZPR2.2 | 17.75 ± 13.61 | 2.07 ± 1.45 | 0.0643 |
| POPTR_0006s08320 | Pt-ZPR3.1 * | 18.88 ± 10.63 | 5.15 ± 1.65 | 0.0467 |
| POPTR_0010s24410 | Pt-ZPR3.2 * | 0.47 ± 0.19 | 2.62 ± 0.80 | 0.0066 |
| Gene name | Leaf blade | Leaf petiole | ||||
|---|---|---|---|---|---|---|
| Expression difference | Mean Cq value in black cottonwood | Mean Cq value in aspen | Expression difference | Mean Cq value in black cottonwood | Mean Cq value in aspen | |
| Pt-AE7.2 | - * | 32.28 | 34.69 | - * | 31.62 | 34.11 |
| Pt-AS1.1 | NS | 35.01 | 34.54 | - * | 32.15 | 34.44 |
| Pt-AS1.2 | + * | 32.92 | 32.27 | NS | 34.04 | 33.81 |
| Pt-AS2.1 | - * | 31.01 | 36.12 | NS | 36.30 | 35.90 |
| Pt-AS2.2 | NS | 32.10 | 33.80 | NS | 34.10 | 36.37 |
| Pt-ATS.2 | - * | 32.97 | 35.11 | NS | 36.71 | 36.53 |
| TIF5A | NS | 28.33 | 28.97 | NS | 27.07 | 28.39 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nowak, J.S.; Douglas, C.J.; Cronk, Q.C.B. Abaxial Greening Phenotype in Hybrid Aspen. Plants 2013, 2, 279-301. https://doi.org/10.3390/plants2020279
Nowak JS, Douglas CJ, Cronk QCB. Abaxial Greening Phenotype in Hybrid Aspen. Plants. 2013; 2(2):279-301. https://doi.org/10.3390/plants2020279
Chicago/Turabian StyleNowak, Julia S., Carl J. Douglas, and Quentin C.B. Cronk. 2013. "Abaxial Greening Phenotype in Hybrid Aspen" Plants 2, no. 2: 279-301. https://doi.org/10.3390/plants2020279




