SPL8 Acts Together with the Brassinosteroid-Signaling Component BIM1 in Controlling Arabidopsis thaliana Male Fertility
Abstract
:1. Introduction
2. Results
2.1. bim1 Enhances the spl8 Semi-Sterile Phenotype

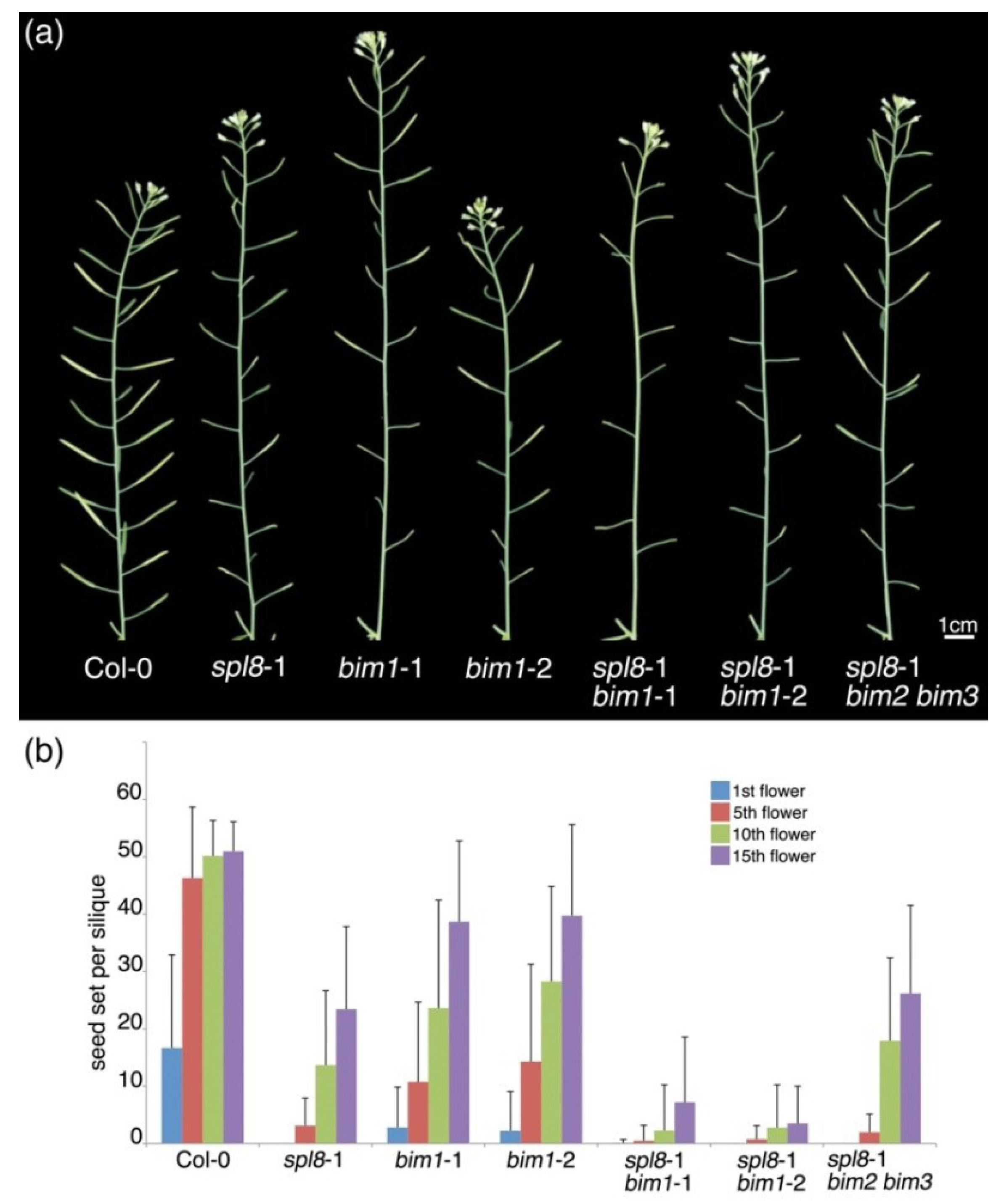
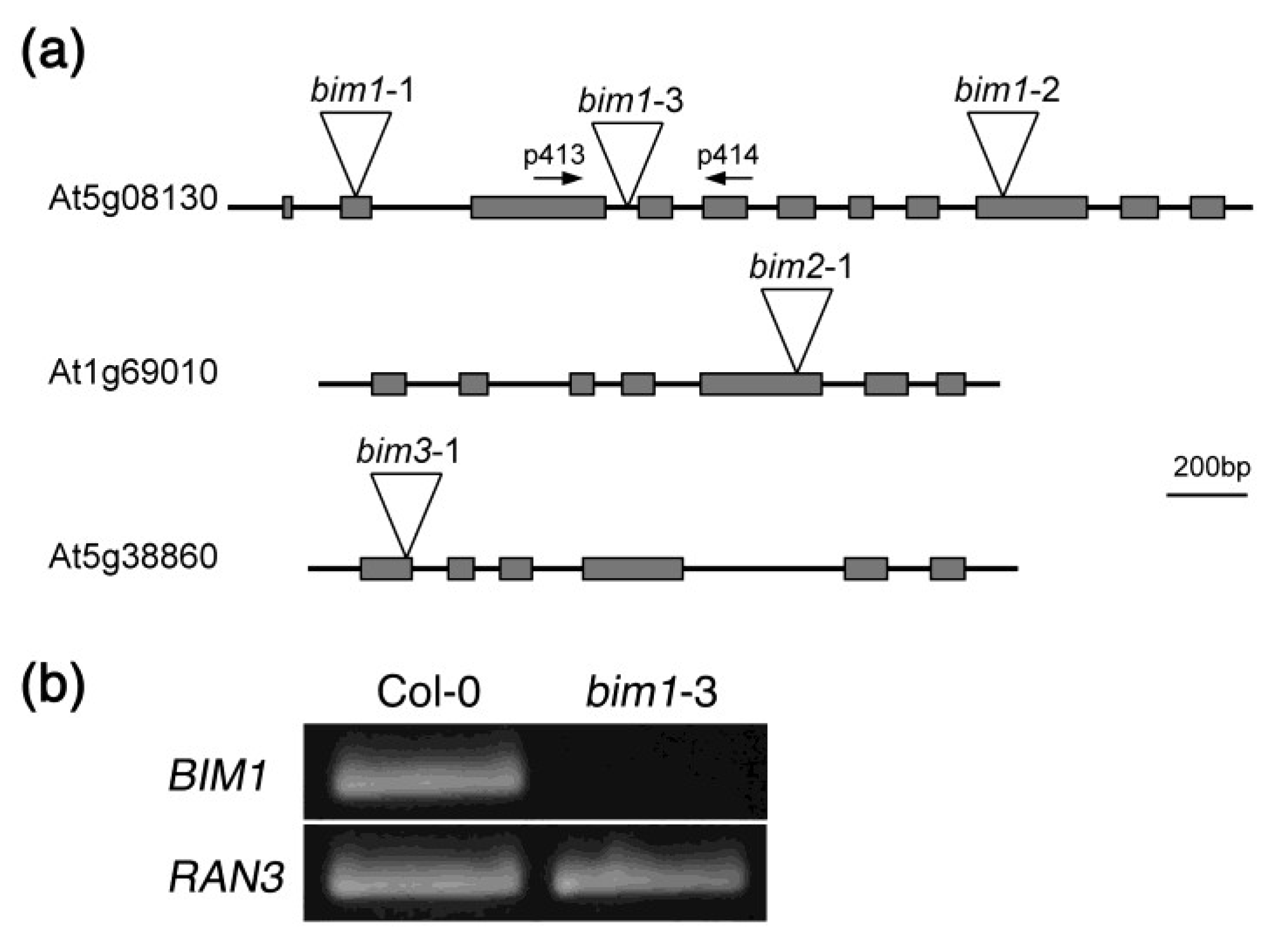
2.2. Isolation of a New Loss-of-Function Allele for BIM1
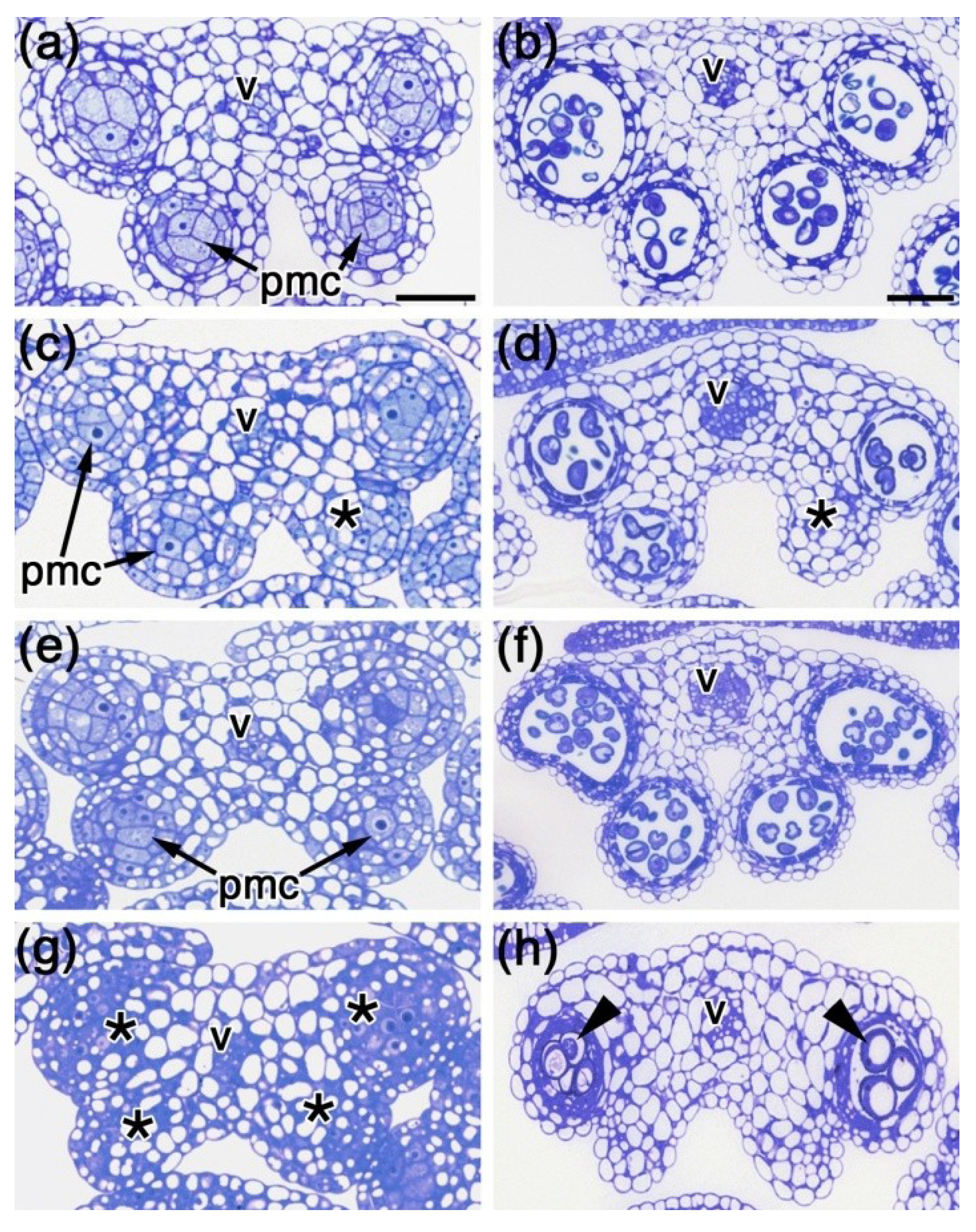
2.3. Histological Analysis of spl8 bim1 Double Mutant Anthers
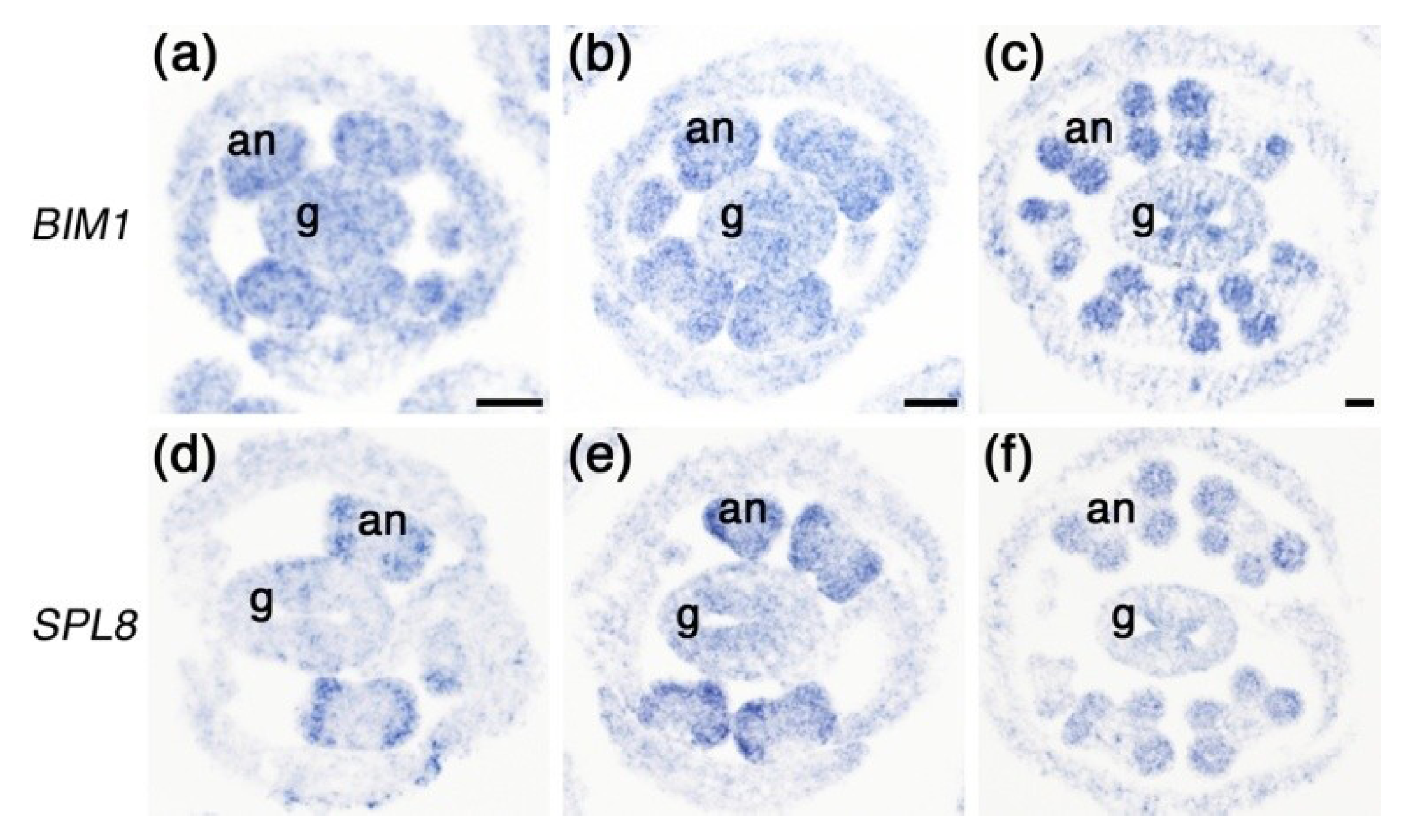
2.4. Expression of BIM1 in Anthers
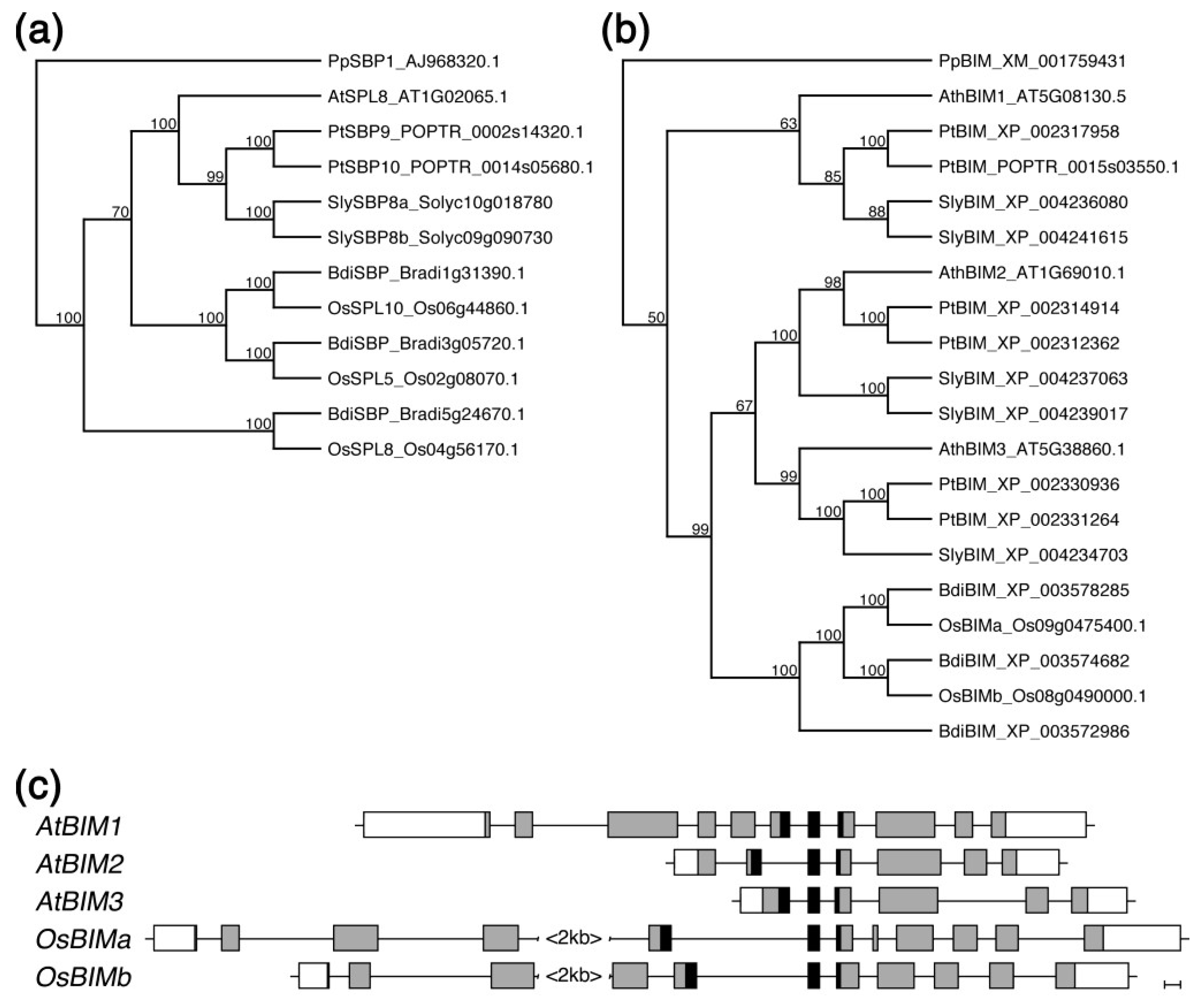
2.5. Both SPL8 and BIM1 Are Conserved in Dicot and Monocot Plants
3. Discussion
4. Experimental Section
4.1. Plant Materials and Growth Conditions
4.2. Histology and Microscopy
4.3. Semi-Quantitative RT-PCR
4.4. In Situ RNA Hybridization
4.5. Phylogenetic Tree Construction
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Bowman, J.L.; Baum, S.F.; Eshed, Y.; Putterill, J.; Alvarez, J. Molecular genetics of gynoecium development in Arabidopsis. Curr. Top. Dev. Biol. 1999, 45, 155–205. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Dickinson, H.G. Stamen structure and function. Plant Cell 2004, 16, S46–S60. [Google Scholar] [CrossRef]
- Causier, B.; Schwarz-Sommer, Z.; Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010, 21, 73–79. [Google Scholar] [CrossRef]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Huijser, P. New players unveiled in early anther development. Plant Signal. Behav. 2011, 6, 934–938. [Google Scholar] [CrossRef]
- Yang, W.C.; Ye, D.; Xu, J.; Sundaresan, V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999, 13, 2108–2117. [Google Scholar] [CrossRef]
- Schiefthaler, U.; Balasubramanian, S.; Sieber, P.; Chevalier, D.; Wisman, E.; Schneitz, K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 11664–11669. [Google Scholar] [CrossRef]
- Xing, S.; Zachgo, S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2008, 53, 790–801. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef]
- Kutschera, U.; Wang, Z.Y. Brassinosteroid action in flowering plants: A darwinian perspective. J. Exp. Bot. 2012, 63, 3511–3522. [Google Scholar] [CrossRef]
- Ye, Q.; Zhu, W.; Li, L.; Zhang, S.; Yin, Y.; Ma, H.; Wang, X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 2010, 107, 6100–6105. [Google Scholar]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- Chandler, J.W.; Cole, M.; Flier, A.; Werr, W. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNRÖSCHEN and DORNRÖSCHEN-LIKE. Plant Mol. Biol. 2009, 69, 57–68. [Google Scholar] [CrossRef]
- Riese, M.; Höhmann, S.; Saedler, H.; Münster, T.; Huijser, P. Comparative analysis of the SBP-Box gene families in P. patens and seed plants. Gene 2007, 401, 28–37. [Google Scholar] [CrossRef]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, J.P.; Tang, L.; Zhao, Y.; Gu, X.C.; Gao, G.; Luo, J.C. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011, 39, D1114–D1117. [Google Scholar] [CrossRef]
- Zhang, Y. The SBP-box gene SPL8 affects reproductive development and gibberellin response in Arabidopsis. Ph.D. Thesis, University of Cologne, Cologne, Germany, 2005. [Google Scholar]
- Cecchetti, V.; Altamur, M.M.; Falasca, G.; Costantino, P.; Cardarelli, M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 2008, 20, 1760–1774. [Google Scholar] [CrossRef]
- Vert, G.; Walcher, C.L.; Chory, J.; Nemhauser, J.L. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. USA 2008, 105, 9829–9834. [Google Scholar]
- Sundberg, E.; Østergaard, L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. 2009, 1, a001628. [Google Scholar] [CrossRef]
- Kirch, T.; Simon, R.; Grünewald, M.; Werr, W. The DORNRÖSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 2003, 15, 694–705. [Google Scholar] [CrossRef]
- Chandler, J.W.; Cole, M.; Flier, A.; Grewe, B.; Werr, W. The AP2 transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 2007, 134, 1653–1662. [Google Scholar] [CrossRef]
- Xing, S.; Rosso, M.G.; Zachgo, S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 2005, 132, 1555–1565. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Xing, S.; Quodt, V.; Chandler, J.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 Acts Together with the Brassinosteroid-Signaling Component BIM1 in Controlling Arabidopsis thaliana Male Fertility. Plants 2013, 2, 416-428. https://doi.org/10.3390/plants2030416
Xing S, Quodt V, Chandler J, Höhmann S, Berndtgen R, Huijser P. SPL8 Acts Together with the Brassinosteroid-Signaling Component BIM1 in Controlling Arabidopsis thaliana Male Fertility. Plants. 2013; 2(3):416-428. https://doi.org/10.3390/plants2030416
Chicago/Turabian StyleXing, Shuping, Vanessa Quodt, John Chandler, Susanne Höhmann, Rita Berndtgen, and Peter Huijser. 2013. "SPL8 Acts Together with the Brassinosteroid-Signaling Component BIM1 in Controlling Arabidopsis thaliana Male Fertility" Plants 2, no. 3: 416-428. https://doi.org/10.3390/plants2030416




