A High-Throughput RNA Extraction for Sprouted Single-Seed Barley (Hordeum vulgare L.) Rich in Polysaccharides
Abstract
:1. Introduction
2. Results and Discussion
3. Conclusions
4. Experimental Section
4.1. Preparation and Buffers
4.2. Buffers (Previously Published in Li and Trick [11])
4.3. Modified from Li and Trick [11] Protocol*
- (1)
- A single 3- to 4-day sprouted barley or oat seed was added to a 1.5 mL microcentrifuge tube with 400 μL of extraction buffer 1 and ground in solution with an RNase sterile micropestle.
- (2)
- An amount of 250 μL of a phenol-chloroform mixture (1:1, pH 4.7) was added and centrifuged immediately at 21,000× g for 30 min at 4 °C.
- (3)
- Approximately 250 μL of the top phase was transferred to a new tube.
- (4)
- An equal volume of extraction buffer 2 was added, the tube was inverted to mix and incubated for 10 min at room temperature.
- (5)
- An amount of 200 μL chloroform-isoamyl alcohol (24:1) was added and centrifuged as above.
- (6)
- The supernatant was removed and added to 300 μL isopropanol and 250 μL 1.2 M sodium chloride in a new tube.
- (7)
- The solution was mixed by inversion and placed on ice for 15 min, then centrifuged at 21,000× g for 30 min at 4 °C.
- (8)
- The supernatant was decanted and the RNA pellet was carefully washed with 400 μL cold 70% ethanol.
- (9)
- The RNA pellet was dried for 10–15 min in a laminar flow hood.
- (10)
- The RNA was dissolved in 50 μL of nuclease-free water and used in downstream applications.
4.4. RNA Analysis and Preparation
4.5. cDNA Synthesis and qRT-PCR
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brookes, P.A.; Lovett, D.A.; MacWilliam, I.C. The steeping of barley: A review of the metabolic consequences of water uptake and their practical implications. J. Inst. Brew. 1976, 82, 14–26. [Google Scholar] [CrossRef]
- Palmer, G.H. Influence of cell wall structure in enzymic breakdown of the endosperm of germinated barley. J. Inst. Brew. 1987, 47, 461–470. [Google Scholar] [CrossRef]
- Potokina, E.; Sreenivasulu, N.; Altschmied, L.; Michalek, W.; Graner, A. Differential gene expression during seed germination in barley (Hordeum vulgare L.). Funct. Integr. Genom. 2002, 2, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Abu-Ghannam, N.; Gallaghar, E. Barley for brewing: Characteristic changes during malting, brewing and applications of its by-products. Comp. Rev. Food Sci. Food Saf. 2010, 9, 318–328. [Google Scholar] [CrossRef]
- Milala, M.A.; Addy, E.O. Hydrolytic enzyme levels in malted cereals. Adv. Biochem. 2014, 2, 76–79. [Google Scholar] [CrossRef]
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination—Still a mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- White, J.; Pacey-Miller, T.; Crawford, A.; Cordeiro, G.; Barbary, D.; Bundock, P.; Henry, R. Abundant transcripts of malting barley identified by serial analysis of gene expression (SAGE). Plant Biotechnol. J. 2006, 4, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Collins, H.M.; Kibble, N.A.; Smith, J.A.; Shirley, N.J.; Jobling, S.A.; Henderson, M.; Singh, R.R.; Pettolino, F.; Wilson, S.M.; et al. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol. J. 2011, 9, 117–135. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, M.; Wright, F.; MacKenzie, K.; Hedley, P.E.; Schwerdt, J.G.; Little, A.; Burton, R.A.; Fincher, G.B.; Marshall, D.; Waugh, R.; et al. The barley genome sequence assembly reveals three additional members of the CslF (1,3;1,4)-β-glucan synthase gene family. PLoS ONE 2014, 9, e90888. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kumar, S.; Singh, P. A quick method to isolate RNA from wheat and other carbohydrate-rich seeds. Plant Mol. Biol. Rep. 2003, 21, 93. [Google Scholar] [CrossRef]
- Li, Z.; Trick, H.N. Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. Biotechniques 2005, 38, 872–875. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, G.; Zhang, X.; Wang, F.; Song, R. Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem. Anal. 2012, 23, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Mornkham, T.; Wangsomnuk, P.P.; Fu, Y.B.; Wangsomnuk, P.; Jogloy, S.; Patanothai, A. Extractions of high quality RNA from the seeds of Jerusalem Artichoke and other plant species with high levels of starch and lipid. Plants 2013, 13, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Luebbehusen, H. The significance of the 260/230 ratio in determining nucleic acid purity. In Genomic and RNA Profiling Core [online]; Baylor College of Medicine: Houston, TX, USA, 2006. [Google Scholar]
- Rashid, A.; Deyholos, M.K. PELPK1 contains a unique pentapeptide repeat and is a positive regulator of germination in Arabidopsis thaliana. Plant Cell Rep. 2011, 30, 1735–1745. [Google Scholar] [CrossRef] [PubMed]

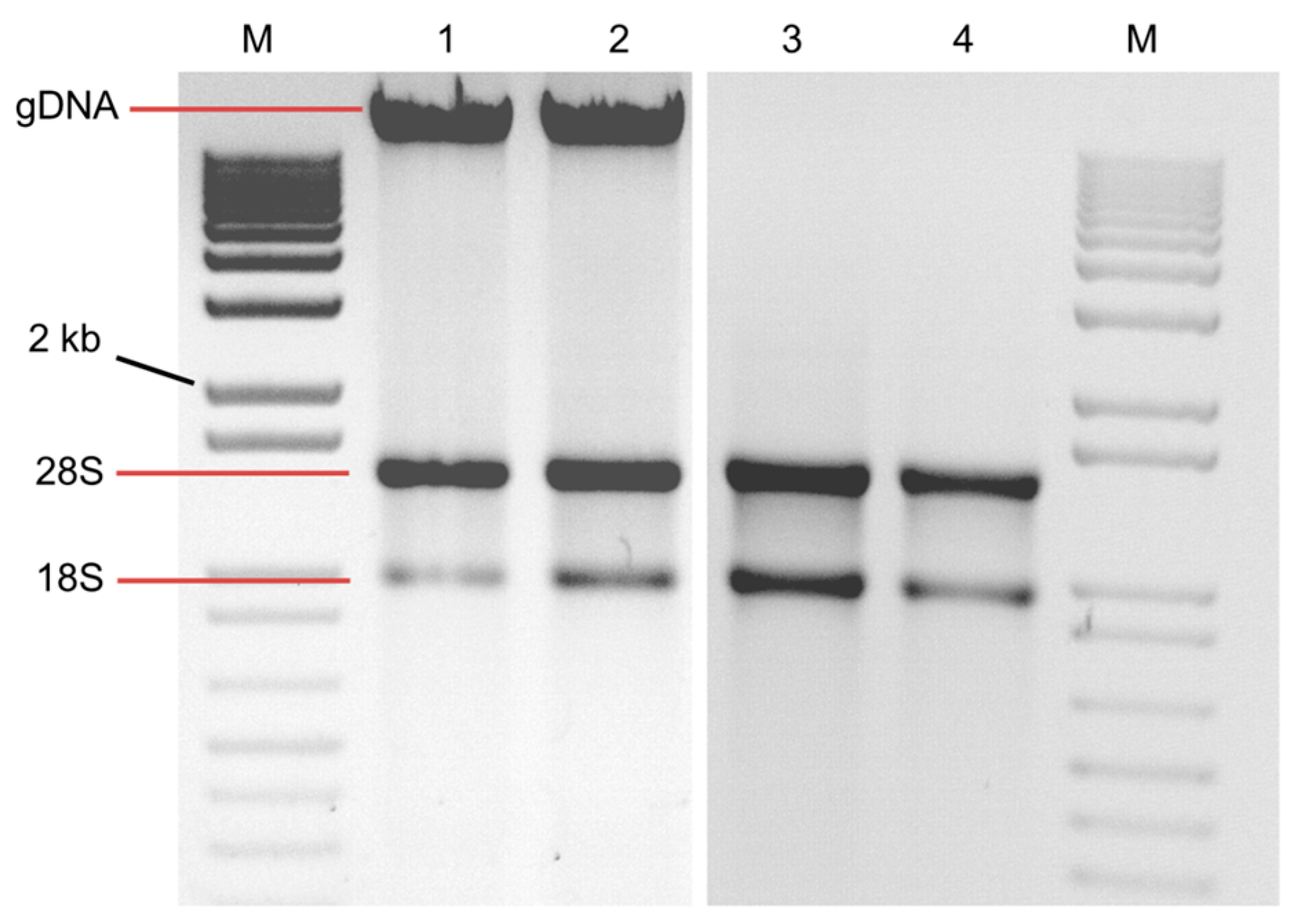
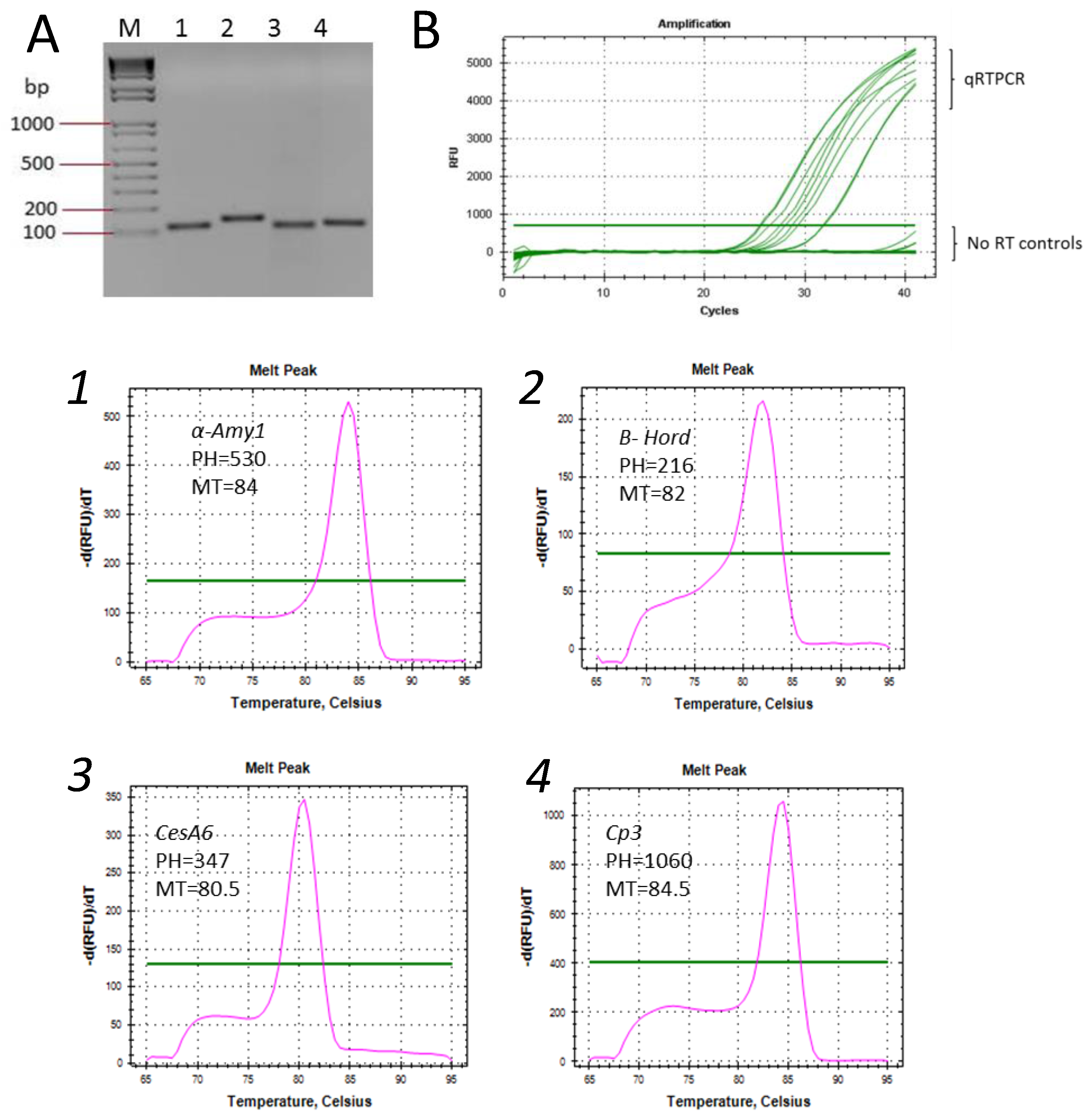
| Absorbance Ratios | |||
|---|---|---|---|
| A260/A280 * | A260/A230 * | ||
| Extraction method | Before–After DNase I Treatment | Before–After DNase I Treatment | RNA per Seed μg/48 mg * |
| GeneJet Column | N/A | N/A | 0.0 |
| TRIzol method (standard) | 1.85c–1.87b | 0.57d–0.61c | 1.1b |
| Li and Trick [11] (standard) | 1.90bc–2.10a | 0.80c–2.20a | 12.9a |
| Li and Trick [11] (modified) | 2.06a–2.05a | 1.32a–2.12a | 14.2a |
| Wang et al. [12] (modified) | 1.99ab–2.08a | 1.05b–1.86b | 14.6a |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, A.; Baldwin, T.; Gines, M.; Bregitzer, P.; Esvelt Klos, K. A High-Throughput RNA Extraction for Sprouted Single-Seed Barley (Hordeum vulgare L.) Rich in Polysaccharides. Plants 2017, 6, 1. https://doi.org/10.3390/plants6010001
Rashid A, Baldwin T, Gines M, Bregitzer P, Esvelt Klos K. A High-Throughput RNA Extraction for Sprouted Single-Seed Barley (Hordeum vulgare L.) Rich in Polysaccharides. Plants. 2017; 6(1):1. https://doi.org/10.3390/plants6010001
Chicago/Turabian StyleRashid, Abdur, Thomas Baldwin, Michael Gines, Phil Bregitzer, and Kathy Esvelt Klos. 2017. "A High-Throughput RNA Extraction for Sprouted Single-Seed Barley (Hordeum vulgare L.) Rich in Polysaccharides" Plants 6, no. 1: 1. https://doi.org/10.3390/plants6010001





