Climate Modelling Shows Increased Risk to Eucalyptus sideroxylon on the Eastern Coast of Australia Compared to Eucalyptus albens
Abstract
:1. Introduction
- Whether the use of solely climate variables is sufficient for projecting future distributions across large areas [31];
- Arbitrariness in the selection of particular nonclimatic variables, and the number of these chosen, and when these are included in the SDM approach, making comparative analyses with similar studies under other approaches difficult [32]. However, a proposal has been made that the adoption of a standard basic configuration of valid ecological variables would enable the testing of SDM ecological assumptions, comparisons with other sets of predictors for similar biophysical processes, as well as providing a basis for the choice of the scale of resolution [33];
- Rating model performance with an independent set of validation data;
- Whether validation by means of an independent set of distribution data is a viable alternative for accuracy of independent data;
- Which techniques consistently outperform others; and
- Whether native, exotic or a combination of both records produces greater modelling efficiency.
2. Methodology
2.1. Distribution Records and Species Phenomena
2.2. Species Distribution Modelling
2.2.1. GLM
2.2.2. MaxEnt
2.2.3. RF
2.2.4. BRT
2.3. Bioclim Variables, Background Data, Test Points & True Skill Statistic (TSS)
3. Results
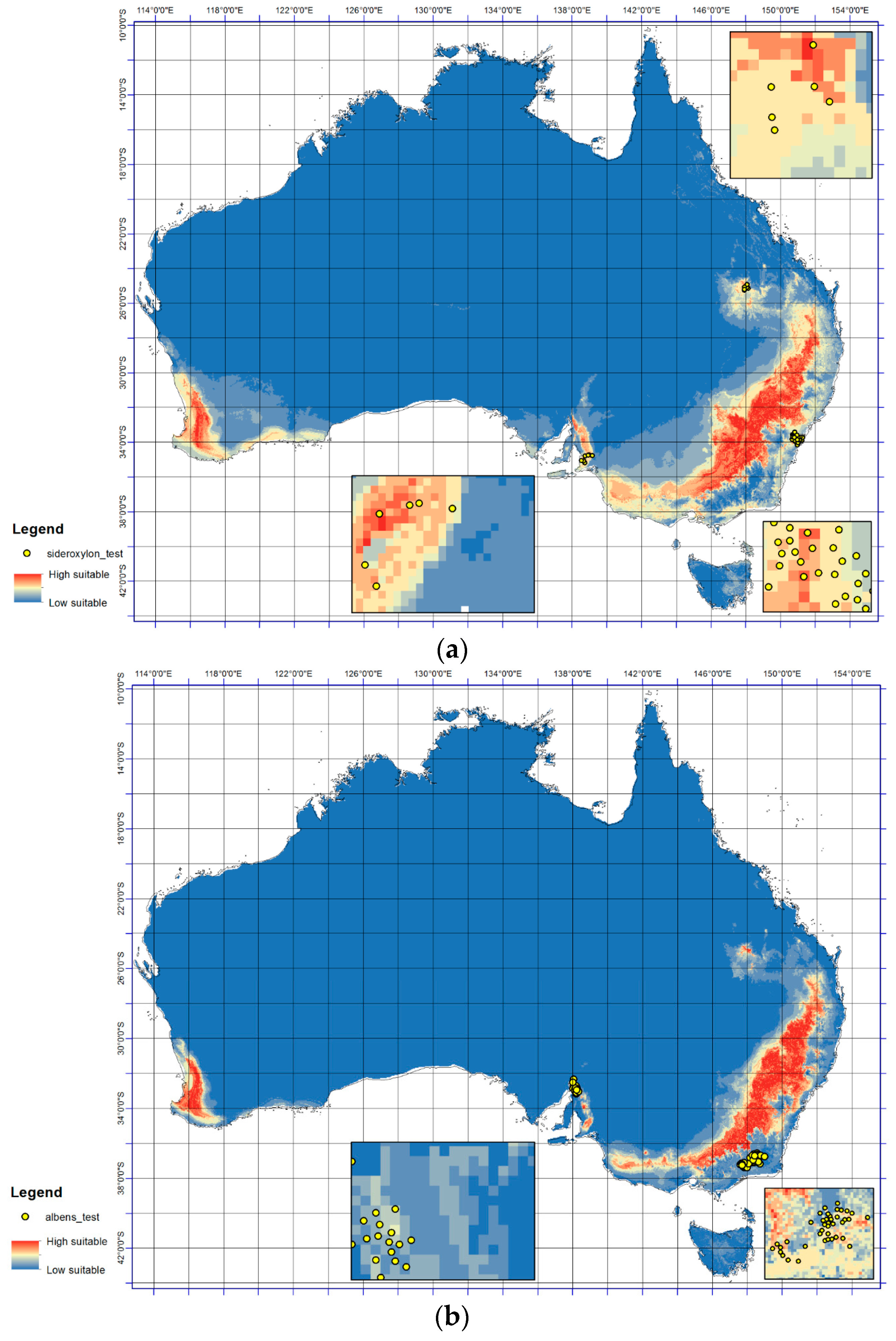
3.1. Model Validation & Models of Current Distribution
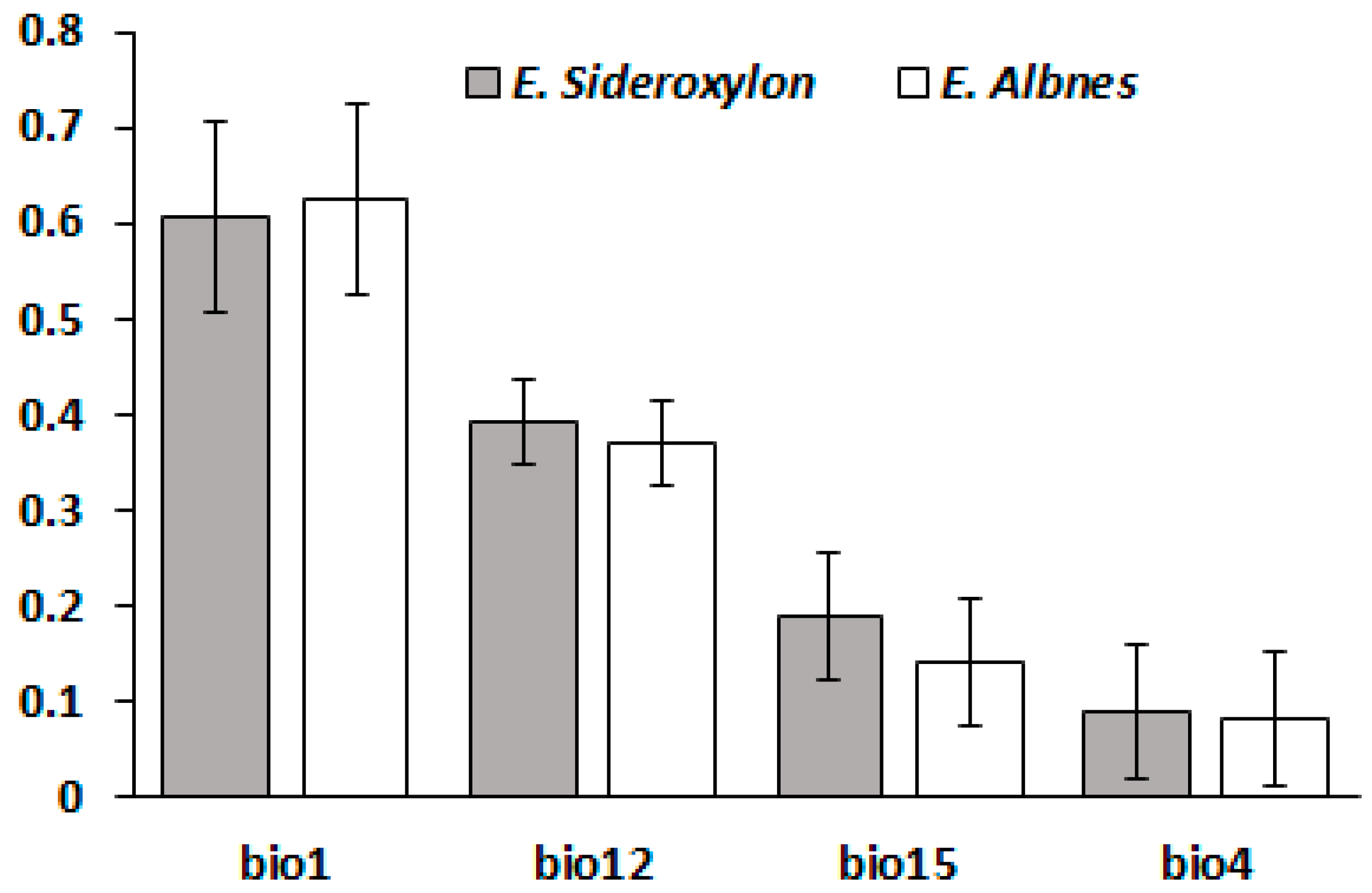
3.2. Variables Contributions on Response Surfaces
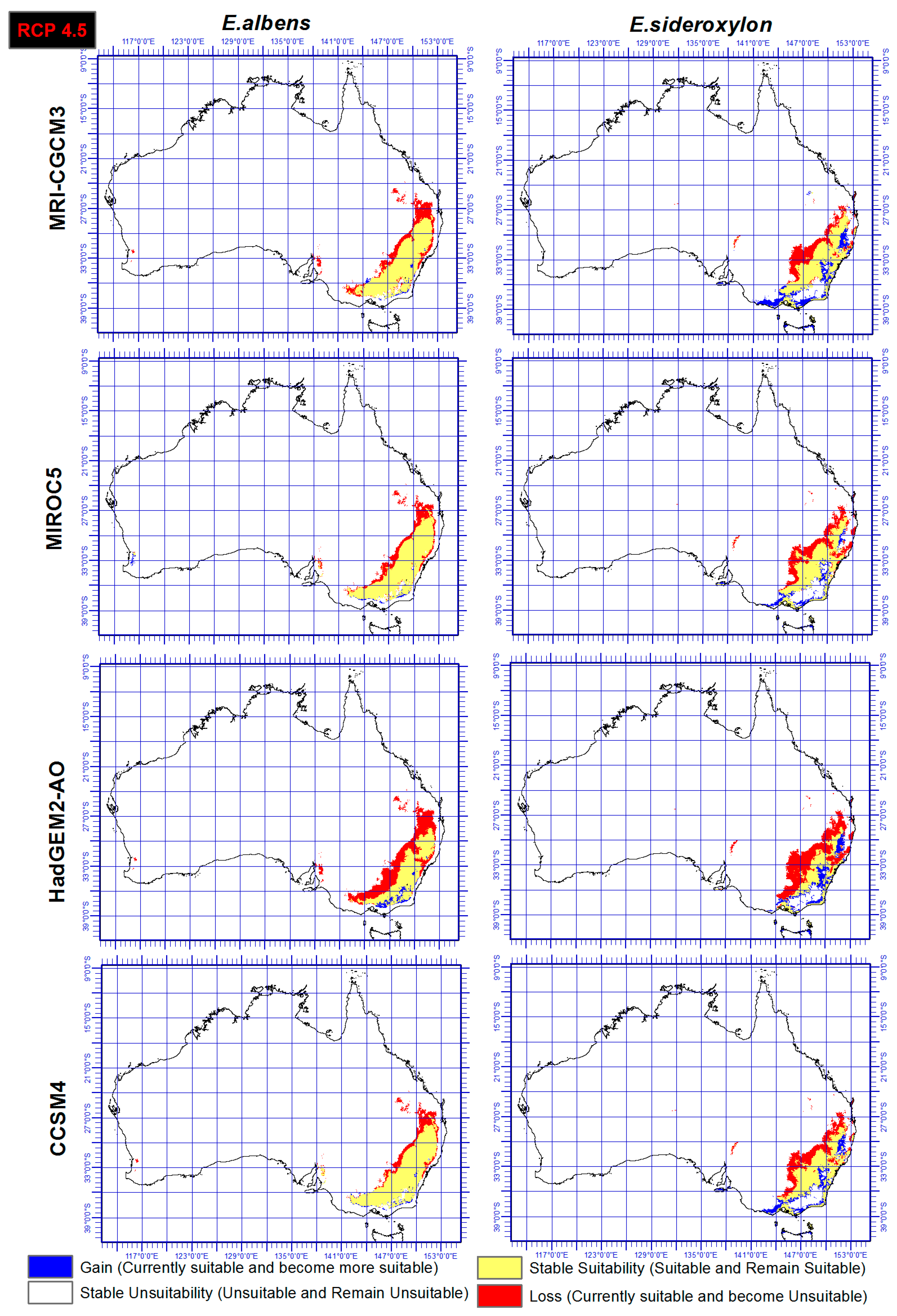
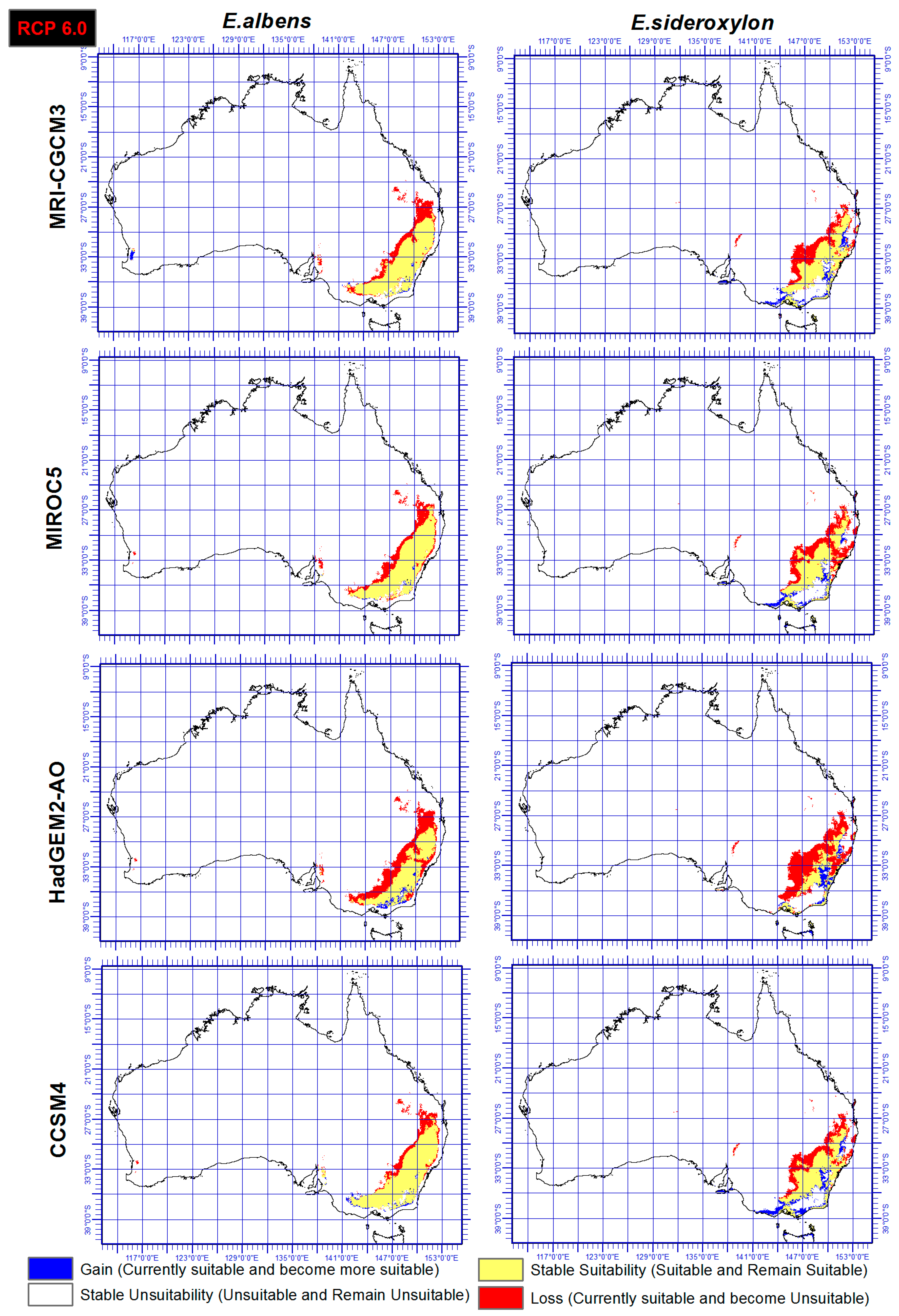
3.3. Range Gain, Loss or As Is
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Australia National Parks and Wildlife Service. White Box-Yellow Box-Blakely’s Red Gum (Box-Gum) Woodland Fact-Sheet; New South Wales Parks and Wildlife Service: Sydney, Australia, 2002.
- Spooner, P.; Allcock, K. Using a state-and-transition approach to manage endangered Eucalyptus albens (White Box) woodlands. Environ. Manag. 2006, 38, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Office of Environment and Heritage. Planting to Conserve Threatened Nomadic Pollinators in NSW; Office of Environment and Heritage: Sydney, Australia, 2016; pp. 12–16.
- Thomas, C.; Cameron, A.; Green, R.; Bakkenes, M.; Beaumont, L.; Collingham, Y.; Erasmus, B.; De Siqueira, M.; Grainger, A.; Hannah, L. Extinction risk from climate change. Nature 2004, 427, 145–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantyka-pringle, C.; Martin, T.; Rhodes, J. Interactions between climate and habitat loss effects on biodiversity: A systematic review and meta-analysis. Glob. Chang. Biol. 2012, 18, 1239–1252. [Google Scholar] [CrossRef]
- Pearson, R.; Dawson, T. Predicting the impacts of climate change on the distribution of species: Are bioclimate envelope models useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N. Predictive habitat distribution models in ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Taylor, S. Climate change impacts on the future distribution of date palms: A modeling exercise using CLIMEX. PLoS ONE 2012, 7, e48021. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-cabral, N.; Kumar, L.; Shabani, F. Future climate scenarios project a decrease in the risk of fall armyworm outbreaks. J. Agric. Sci. 2017, 1–20. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Ahmadi, M.; Esmaeili, A. Are research efforts on Animalia in the South Pacific associated with the conservation status or population trends? J. Nat. Conserv. 2017. [Google Scholar] [CrossRef]
- Da Silva, R.; Kumar, L.; Shabani, F.; Picanço, M. Potential risk levels of invasive Neoleucinodes elegantalis (small tomato borer) in areas optimal for open-field Solanum lycopersicum (tomato) cultivation in the present and under predicted climate change. Pest Manag. Sci. 2017, 73, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Shabani, F.; Kumar, L.; Nojoumian, A.; Esmaeili, A.; Toghyani, M. Projected future distribution of date palm and its potential use in alleviating micronutrient deficiency. J. Sci. Food Agric. 2016, 96, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.; Kumar, L.; Shabani, F.; da Silva, E.; da Silva Galdino, T.; Picanço, M. Spatio-temporal dynamic climate model for Neoleucinodes elegantalis using CLIMEX. Int. J. Biometeorol. 2017, 61, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Thornton, K. The potential impacts of climate change on maize production in Africa and Latin America in 2055. Glob. Environ. Chang. 2003, 13, 51–59. [Google Scholar] [CrossRef]
- Reddy, K.R.; Hodges, H.F.; McKinion, J.M. A comparison of scenarios for the effect of global climate change on cotton growth and yield. Funct. Plant Biol. 1997, 24, 707–713. [Google Scholar] [CrossRef]
- Rogers, D.J.; Reid, R.E.; Rogers, J.J.; Addison, S.J. Prediction of the naturalisation potential and weediness risk of transgenic cotton in Australia. Agric. Ecosyst. Environ. 2007, 119, 177–189. [Google Scholar] [CrossRef]
- Shabani, F.; Kotey, B. Future distribution of cotton and wheat in Australia under potential climate change. J. Agric. Sci. 2015, 1–11. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Esmaeili, A. Future distributions of Fusarium oxysporum f. spp. in European, Middle Eastern and North African agricultural regions under climate change. Agric. Ecosyst. Environ. 2014, 197, 96–105. [Google Scholar] [CrossRef]
- Ortiz, R.; Sayre, K.D.; Govaerts, B.; Gupta, R.; Subbarao, G.; Ban, T.; Hodson, D.; Dixon, J.M.; Iván Ortiz-Monasterio, J.; Reynolds, M. Climate change: Can wheat beat the heat? Agric. Ecosyst. Environ. 2008, 126, 46–58. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Taylor, S. Projecting date palm distribution in Iran under climate change using topography, physicochemical soil properties, soil taxonomy, land use and climate data. Theor. Appl. Climatol. 2014, 152, 543–557. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Esmaeili, A.; Saremi, H. Climate change will lead to larger areas of Spain being conducive to date palm cultivation. J. Food Agric. Environ. 2013, 11, 2441–2446. [Google Scholar]
- Pearson, R.G.; Dawson, T.P.; Berry, P.M.; Harrison, P.A. SPECIES: A spatial evaluation of climate impact on the envelope of species. Ecol. Model. 2002, 154, 289–300. [Google Scholar] [CrossRef]
- Pattison, R.R.; Mack, R.N. Potential distribution of the invasive tree Triadica sebifera (Euphorbiaceae) in the United States: Evaluating CLIMEX predictions with field trials. Glob. Chang. Biol. 2008, 14, 813–826. [Google Scholar] [CrossRef]
- Paterson, R.; Kumar, L.; Shabani, F.; Lima, N. World climate suitability projections to 2050 and 2100 for growing oil palm. J. Agric. Sci. 2017, 155, 689–702. [Google Scholar] [CrossRef]
- Lamsal, P.; Kumar, L.; Shabani, F.; Atreya, K. The greening of the Himalayas and Tibetan Plateau under climate change. Glob. Planet. Chang. 2017. [Google Scholar] [CrossRef]
- Ramirez-Cabral, N.; Kumar, L.; Shabani, F. Global risk levels for corn rusts (Puccinia sorghi and Puccinia polysora) under climate change projections. J. Phytopathol. 2017, 165, 563–574. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Esmaeili, A. A modelling implementation of climate change on biodegradation of Low-Density Polyethylene (LDPE) by Aspergillus niger in soil. Glob. Ecol. Conserv. 2015, 4, 388–398. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Willis, K.; Bhagwat, S. Biodiversity and climate change. Science 2009, 326, 806–807. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, J.M.; Strayer, D.L. Usefulness of bioclimatic models for studying climate change and invasive species. Ann. N. Y. Acad. Sci. 2008, 1134, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Araujo, M.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Randin, C.; Engler, R.; Normand, S.; Zappa, M.; Zimmermann, N.; Pearman, P.; Vittoz, P.; Thuiller, W.; Guisan, A. Climate change and plant distribution: Local models predict high-elevation persistence. Glob. Chang. Biol. 2009, 15, 1557–1569. [Google Scholar] [CrossRef]
- Austin, M.; Van Niel, K. Improving species distribution models for climate change studies: Variable selection and scale. J. Biogeogr. 2011, 38, 1–8. [Google Scholar] [CrossRef]
- Global Biodiversity Information Facility. Global Biodiversity Information Facility (GBIF). Available online: http://www.gbif.org (accessed on 25 July 2015).
- Atlas of Living Australia. Available online: http://www.ala.org.au/ (accessed on 25 July 2017).
- Dormann, C.; McPherson, J.; Araújo, M.; Bivand, R.; Bolliger, J.; Carl, G.; Davies, R.G.; Hirzel, A.; Jetz, W.; Daniel Kissling, W. Methods to account for spatial autocorrelation in the analysis of species distributional data: A review. Ecography 2007, 30, 609–628. [Google Scholar] [CrossRef]
- Ahmadi, M.; Nezami Balouchi, B.; Jowkar, H.; Hemami, M.; Fadakar, D.; Malakouti-Khah, S.; Ostrowski, S. Combining landscape suitability and habitat connectivity to conserve the last surviving population of cheetah in Asia. Divers. Distrib. 2017, 23, 592–603. [Google Scholar] [CrossRef]
- Warren, D.; Glor, R.; Turelli, M. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- International Union for Conservation of Nature. Available online: http://www.iucn.org/ (accessed on 25 July 2017).
- Boland, D.; Brooker, M.; Chippendale, G.; Hall, N.; Hyland, B.; Johnston, R.; Kleinig, D.; McDonald, M.; Turner, J. Forest Trees of Australia; CSIRO Publishing: Clayton, Australia, 2006. [Google Scholar]
- McCullagh, P.; Nelder, J. Generalized Linear Models. In Standard Book on Generalized Linear Models, 2nd ed.; Chapman-Hall: London, UK, 1989. [Google Scholar]
- Phillips, S.; Anderson, R.; Schapire, R. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.; Hastie, T. A working guide to boosted regression trees. J. Anim. Ecol. 2008, 77, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Thuiller, W.; Lafourcade, B.; Engler, R.; Araújo, M. BIOMOD—A platform for ensemble forecasting of species distributions. Ecography 2009, 32, 369–373. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Guillera-Arroita, G.; Lahoz-Monfort, J.; Elith, J. Maxent is not a presence–absence method: A comment on Thibaud et al. Methods Ecol. Evol. 2014, 5, 1192–1197. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.; Hastie, T.; Dudík, M.; Chee, Y.; Yates, C. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Elith, J.; Kearney, M.; Phillips, S. The art of modelling range-shifting species. Methods Ecol. Evol. 2010, 1, 330–342. [Google Scholar] [CrossRef]
- Svetnik, V.; Liaw, A.; Tong, C.; Culberson, J.; Sheridan, R.; Feuston, B. Random forest: A classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003, 43, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Ridgeway, G. Gbm: Generalized Boosted Regression Models. 2006. Available online: https://cran.r-project.org/web/packages/dismo/vignettes/brt.pdf (accessed on 25 July 2017).
- Hijmans, R.; Cameron, S.; Parra, J.; Jones, P.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Shabani, F.; Kumar, L.; Ahmadi, M. A comparison of absolute performance of different correlative and mechanistic species distribution models in an independent area. Ecol. Evol. 2016, 6, 5973–5986. [Google Scholar] [CrossRef] [PubMed]
- Araújo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Randall, D.; Wood, R.; Bony, S.; Colman, R.; Fichefet, T.; Fyfe, J.; Kattsov, V.; Pitman, A.; Shukla, J.; Srinivasan, J. Climate models and their evaluation. In Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the IPCC (FAR); Cambridge University Press: Cambridge, UK, 2007; pp. 589–662. [Google Scholar]
- Ghosh, S.; Mujumdar, P. Climate change impact assessment: Uncertainty modeling with imprecise probability. J. Geophys. Res. Atmos. 2009, 114. [Google Scholar] [CrossRef]
- Weiland, F.; Van Beek, L.; Kwadijk, J.; Bierkens, M. The ability of a GCM-forced hydrological model to reproduce global discharge variability. Hydrol. Earth Syst. Sci. 2010, 14, 1595. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.; Mehrotra, R.; Sharma, A. Can the variability in precipitation simulations across GCMs be reduced through sensible bias correction? Clim. Dyn. 2017, 1–19. [Google Scholar] [CrossRef]
- Cannon, A.; Sobie, S.; Murdock, T. Bias Correction of GCM Precipitation by Quantile Mapping: How Well Do Methods Preserve Changes in Quantiles and Extremes? J. Clim. 2015, 28, 6938–6959. [Google Scholar] [CrossRef]

| TSS (Testing, Training) | ||||
|---|---|---|---|---|
| GLM | MaxEnt | RF | BRT | |
| E. sideroxylon | 0.66, 0.86 | 0.67, 0.88 | 0.64, 0.88 | 0.67, 0.88 |
| E. albens | 0.78, 0.92 | 0.79, 0.92 | 0.76, 0.92 | 0.78, 0.92 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shabani, F.; Kumar, L.; Ahmadi, M. Climate Modelling Shows Increased Risk to Eucalyptus sideroxylon on the Eastern Coast of Australia Compared to Eucalyptus albens. Plants 2017, 6, 58. https://doi.org/10.3390/plants6040058
Shabani F, Kumar L, Ahmadi M. Climate Modelling Shows Increased Risk to Eucalyptus sideroxylon on the Eastern Coast of Australia Compared to Eucalyptus albens. Plants. 2017; 6(4):58. https://doi.org/10.3390/plants6040058
Chicago/Turabian StyleShabani, Farzin, Lalit Kumar, and Mohsen Ahmadi. 2017. "Climate Modelling Shows Increased Risk to Eucalyptus sideroxylon on the Eastern Coast of Australia Compared to Eucalyptus albens" Plants 6, no. 4: 58. https://doi.org/10.3390/plants6040058





