Effect of Aluminum Treatment on Proteomes of Radicles of Seeds Derived from Al-Treated Tomato Plants
Abstract
:1. Introduction
2. Experimental
2.1. Protein Extraction and Isobaric Tags for Relative and Absolute Quantification Labeling
2.2. Data Processing, Database Searching and iTRAQ Quantitation
2.3. Protein Quantification, Statistics, and Protein Functional Analysis
3. Results and Discussion
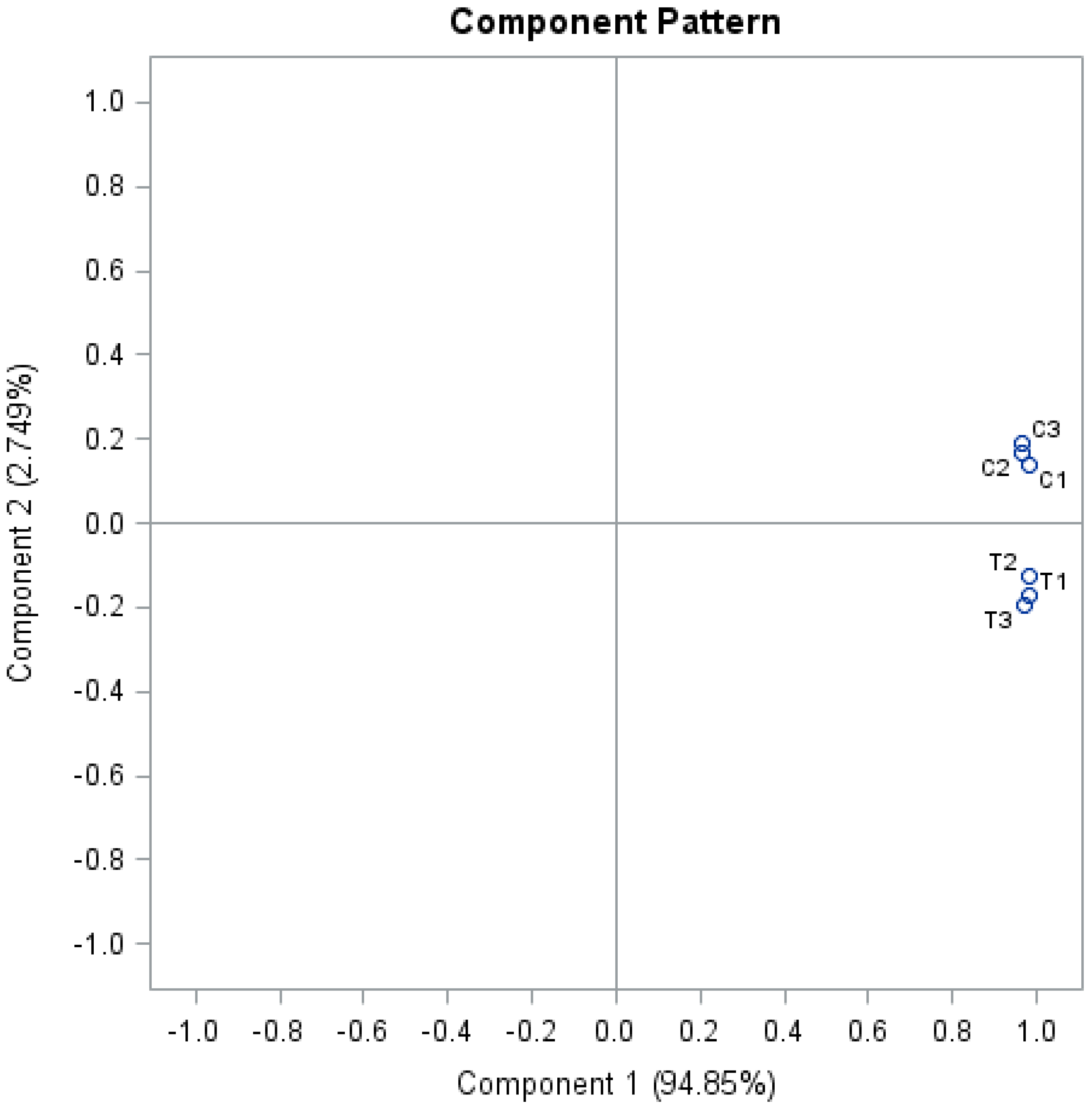
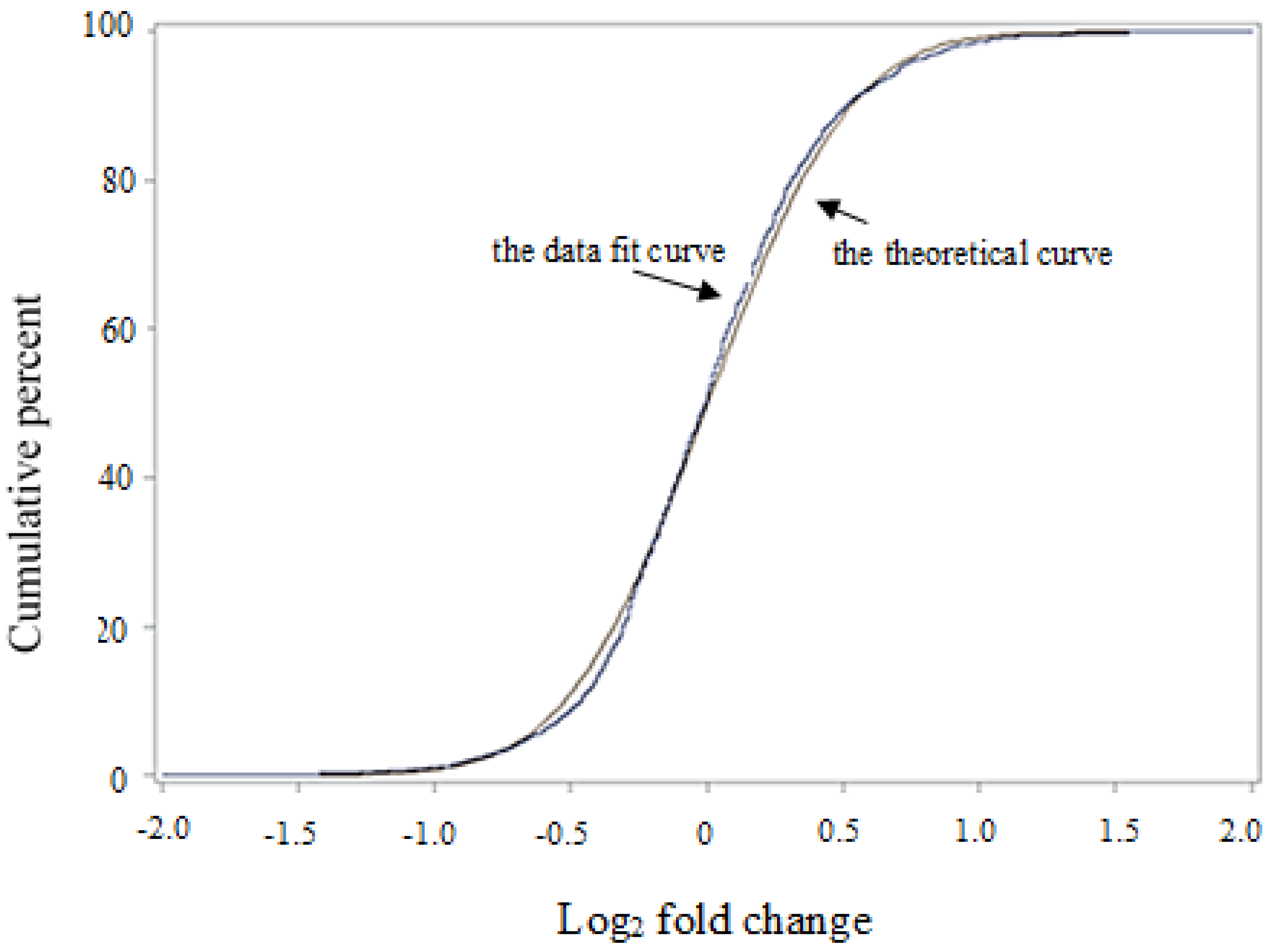
3.1. Al Treatment-Induced Proteome Changes and the Associated Cellular and Molecular Functions
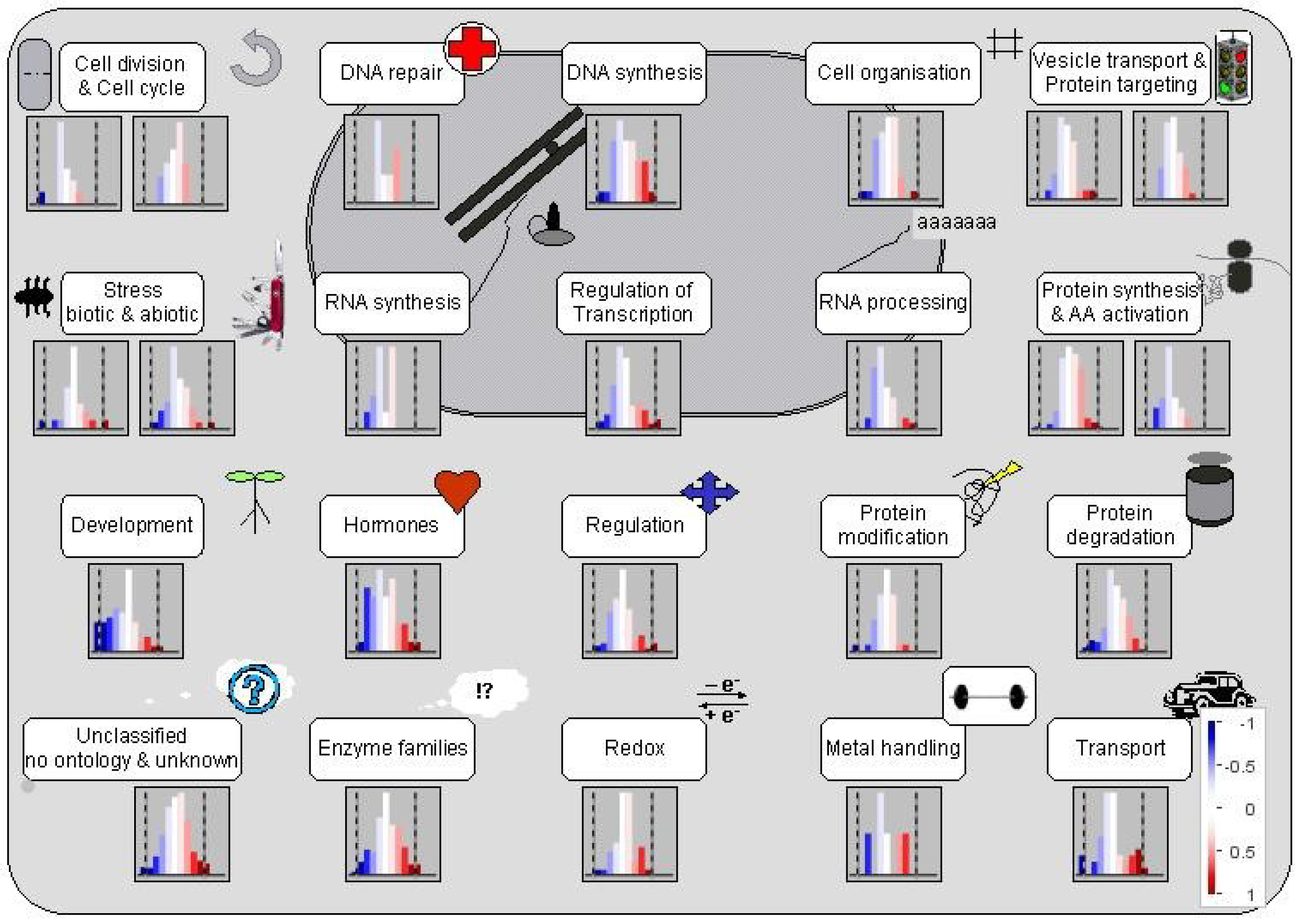
3.1.1. Mobilization of Seed Storage Proteins in Al-Treated Tomato Radicles
3.1.2. Proteins Involved in Cell Organization, Cell Division and Cell Cycle
3.1.3. Proteins Involved in Regulation of Transcription
3.1.4. Proteins Affecting Protein Synthesis and Post-Translational Modification
3.1.5. Proteins Involved in Protein Degradation and Modification
3.1.6. Proteins Involved in Hormone Metabolism and Signaling
3.1.7. Proteins in Signal Transduction
3.1.8. Stress Proteins
3.1.9. Enzymes in Cellular Metabolism
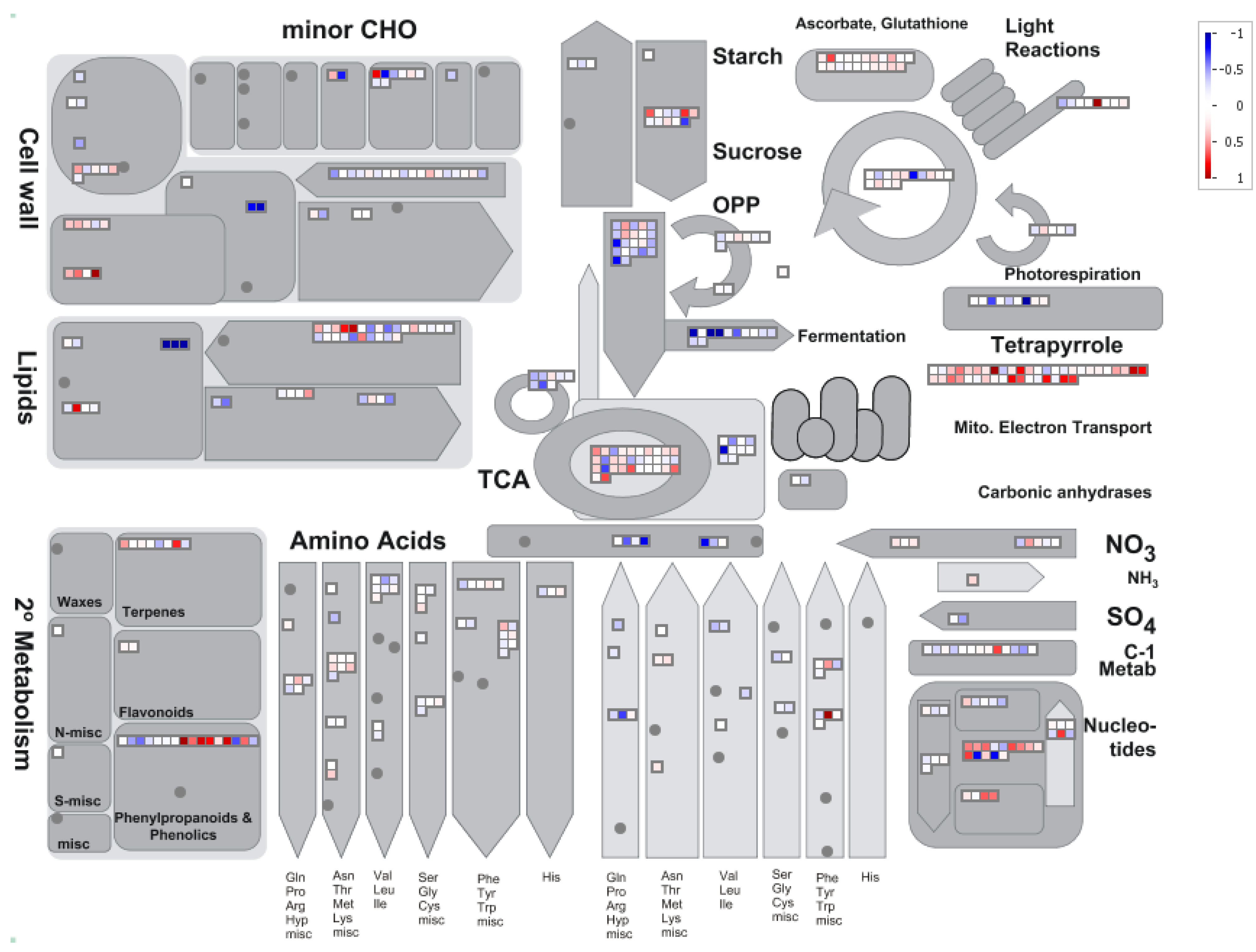
4. Conclusions
- The Al-treated radicles contained lower abundance of hydrophilic (seed) proteins and oil body membrane proteins, which could reduce the tolerance to dehydration. This could cause the tomato seedlings to be more susceptible to drought and salt and other factors;
- Mitochondria function was enhanced because of the active protein translational machinery (induction in ribosomal proteins) and TCA–oxidative phosphorylation cycle;
- The identified proteins include regulatory proteins for gene expression, signaling pathways, cell cycle and programmed cell death.
Acknowledgments
Author Contributions
Conflicts of Interest
References and Notes
- Nasr, N. Germination and seedling growth of maize (Zea mays L.) seeds in toxicity of aluminum and nickel. Merit Res. J. Environ. Sci. Toxic. 2013, 1, 110–113. [Google Scholar]
- Zhu, L.; Wang, J.; Fang, X.; Wang, Y.; Hao, J.; Weiwei, M.; Jiao, T. Effect of seed soaking with aluminum on seed germination and seedling physiology of Platycodon grandiflorum. Zhongguo Zhong Yao Za Zhi 2010, 35, 3255–3259. [Google Scholar]
- Liang, C.; Piñeros, M.A.; Tian, J.; Yao, Z.F.; Sun, L.L.; Liu, J.P.; Shaff, J.; Coluccio, A.; Kochian, L.V.; Liao, H. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013, 161, 1347–1361. [Google Scholar] [CrossRef]
- Liao, H.; Wan, H.; Shaff, J.; Wang, X.; Yan, X.; Kochian, L.V. Phosphorus and aluminuminteractions in soybean in relation to aluminum tolerance: Exudation of specific organic acidsfrom different regions of the intact root system. Plant Physiol. 2006, 141, 674–684. [Google Scholar] [CrossRef]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Piñeros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef]
- Yang, L.T.; Qi, Y.P.; Jiang, H.X.; Chen, L.S. Roles of organic acid anion secretion in aluminum tolerance of higher plants. Biomed. Res. Int. 2013, 2013, e173682. [Google Scholar] [CrossRef]
- Eticha, D.; Stass, A.; Horst, W.J. Cell-wall pectin and its degree of methylation in the maize root-apex: Significance for genotypic differences in aluminium resistance. Plant Cell. Environ. 2005, 28, 1410–1420. [Google Scholar] [CrossRef]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar]
- Huang, C.F.; Yamaji, N.; Chen, Z.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012, 69, 857–867. [Google Scholar] [CrossRef]
- Zhu, X.F.; Lei, G.J.; Wang, Z.W.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Coordination between apoplastic and symplastic detoxification confers plant aluminum resistance. Plant Physiol. 2013, 162, 1947–1955. [Google Scholar] [CrossRef]
- Bewley, J.D.; Black, M. Seeds, Physiology of Development and Germination, 2nd ed.; Plenum Press: New York, NY, USA, 1994. [Google Scholar]
- Bewley, J.D. Seed germination and Dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef]
- Koornneef, M.; Bentsink, L.; Karssen, C.M. Seed dormancy and germination. Curr. Opin. Plant Biol. 2002, 5, 33–36. [Google Scholar] [CrossRef]
- Di Salvatore, M.; Carafa, A.M.; Carratù, G. Assessment of heavy metals phytotoxicity using seed germination and root elongation tests: A comparison of two growth substrates. Chemosphere 2008, 73, 1461–1464. [Google Scholar] [CrossRef]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef]
- Aniol, A. Induction of aluminum tolerance in wheat seedlings by low doses of aluminum in the nutrient solution. Plant Physiol. 1984, 76, 551–555. [Google Scholar]
- Zhou, S.P.; Ikenna, O.; Rutzke, M.A.; Giri, S.K. Tennessee State University: Nashville, TN, USA, Unpublished data. 2014.
- Zhou, S.; Palmer, M.; Zhou, J.; Bhatti, S.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Differential root proteome expression in tomato genotypes with contrasting drought tolerance exposed to dehydration. J. Am. Soc. Hortic. Sci. 2013, 138, 1–11. [Google Scholar]
- Thannhauser, T.W.; Shen, M.; Sherwood, R.; Howe, K.; Fish, T.; Yang, Y.; Chen, W.; Zhang, S. A workflow for large-scale empirical identification of cell wall N-linked glycoproteins of tomato (Solanum lycopersicum) fruit by tandem mass spectrometry. Electrophoresis 2013, 34, 2417–2431. [Google Scholar] [CrossRef]
- Yang, Y.; Qiang, X.; Owsiany, K.; Zhang, S.; Thannhauser, T.W.; Li, L. Evaluation of different multidimensional LC-MS/MS pipelines for iTRAQ-based proteomic analysis of potato tubers in response to cold storage. J. Proteome Res. 2011, 10, 4647–4660. [Google Scholar] [CrossRef]
- International Tomato Genome Sequencing Project. Available online: http://solgenomics.net/tomato/ (accessed on 16 September 2011).
- MapMan, version 3.1.0. Available online: http://mapman.gabipd.org/web/guest/mapman (accessed on 30 January 2014).
- Delhaize, E.; Ma, J.F.; Ryan, P.R. Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 2012, 17, 341–348. [Google Scholar] [CrossRef]
- Famoso, A.N.; Zhao, K.; Clark, R.T.; Tung, C.W.; Wright, M.H.; Bustamante, C.; Kochian, L.V.; McCouch, S.R. Genetic Architecture of Aluminum Tolerance in Rice (Oryza sativa) Determined through Genome-Wide Association Analysis and QTL Mapping. PLoS Genet. 2011, 7, e1002221. [Google Scholar] [CrossRef]
- Ryan, P.R.; Tyerman, S.D.; Sasaki, T.; Furuichi, T.; Yamamoto, Y.; Zhang, W.H.; Delhaize, E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 2011, 62, 9–20. [Google Scholar] [CrossRef]
- Zhou, S.; Sauvé, R.; Thannhauser, T.W. Proteome changes induced by aluminium stress in tomato roots. J. Exp. Bot. 2009, 60, 1849–1857. [Google Scholar] [CrossRef]
- Kim, Y.Y.; Jung, K.W.; Yoo, K.S.; Jeung, J.U.; Shin, J.S. A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol. 2011, 52, 874–884. [Google Scholar] [CrossRef]
- Poxleitner, M.; Rogers, S.W.; Samuels, A.L.; Browse, J.; Rogers, J.C. A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 2006, 47, 917–933. [Google Scholar] [CrossRef]
- Murphy, D.J.; Hernandez-Pinzon, I.; Patel, K. Role of lipid bodies and lipid-body proteins in seeds and others tissues. J. Plant Physiol. 2001, 158, 471–478. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F., III; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 22, 743–754. [Google Scholar]
- Levine, A.; Belenghi, B.; Damari-Weisler, H.; Granot, D. Vesicle-associated membrane protein of Arabidopsis suppresses Bax-induced apoptosis in yeast downstream of oxidative burst. J. Biol. Chem. 2001, 276, 46284–46289. [Google Scholar] [CrossRef]
- Harris, E.H.; Boynton, J.E.; Gillham, N.W. Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 1994, 58, 700–754. [Google Scholar]
- Pietromonaco, S.F.; Denslow, N.D.; O'Brien, T.W. Proteins of mammalian mitochondrial ribosomes. Biochimie 1991, 73, 827–835. [Google Scholar] [CrossRef]
- Spremulli, L. Protein synthesis of mammalian mitochondria. Available online: http://www.chem.unc.edu/people/faculty/spremulli/ (accessed on 10 February 2014).
- Zhou, S.; Sauvé, R.J.; Liu, Z.; Reddy, S.; Bhatti, S.; Hucko, S.D.; Fish, T.; Thannhauser, T.W. Identification of salt-induced changes in leaf and root proteomes of the wild tomato, Solanum chilense. J. Am. Soc. Hortic. Sci. 2011, 136, 288–302. [Google Scholar]
- Berndt, N. Roles and regulation of serine/threonine-specific protein phosphatases in the cell cycle. Prog. Cell Cycle Res. 2003, 5, 497–510. [Google Scholar]
- Garcia, A.; Cayla, X.; Guergnon, J.; Dessauge, F.; Hospital, V.; Rebollo, M.P.; Fleischer, A.; Rebollo, A. Serine/threonine protein phosphatases PP1 and PP2A are key players in apoptosis. Biochimie 2003, 85, 721–726. [Google Scholar] [CrossRef]
- Osawa, H.; Matsumoto, H. Possible involvement of protein phosphorylation in aluminum-responsive malate efflux from wheat root apex. Plant Physiol. 2001, 126, 411–420. [Google Scholar] [CrossRef]
- Alcaide-Loridan, C.; Jupin, I. Ubiquitin and Plant Viruses, Let’s Play Together! Plant Physiol. 2012, 160, 72–82. [Google Scholar] [CrossRef]
- Nezames, C.D.; Deng, X.Y. The COP9 Signalosome: Its regulation of Cullin-Based E3 Ubiquitin ligases and role in photomorphogenesis. Plant Physiol. 2012, 160, 38–46. [Google Scholar]
- Wei, N.; Serino, G.; Deng, X.W. The COP9 signalosome: More than a protease. Trends Biochem. Sci. 2008, 33, 592–600. [Google Scholar] [CrossRef]
- Argyris, J.; Dahal, P.; Hayashi, E.; Still, D.W.; Bradford, K.J. Genetic Variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 2008, 148, 926–947. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Lynch, T.J. Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol. 2000, 122, 1179–1186. [Google Scholar] [CrossRef]
- Orton, R.J.; Sturm, O.E.; Vyshemirsky, V.; Calder, M.; Gilbert, D.R.; Kolch, W. Computational modelling of the receptor-tyrosine-kinase-activated MAPK pathway. Biochem. J. 2005, 392, 249–261. [Google Scholar] [CrossRef]
- Arroyo-Serralta, G.A.; Ku´-Gonza´lez, A.; Herna´ndez-Sotomayor, S.M.T.; Zu´n˜iga-Aguilar, J.J. Exposure to toxic concentrations of Aluminum activates a MAPK-like protein in cell suspension cultures of Coffea arabica. Plant Physiol. Biochem. 2005, 43, 27–35. [Google Scholar] [CrossRef]
- Ramı´rez-Benı´tez, J.E.; Chee-Gonza´lez, L.; Herna´ndez-Sotomayor, S.M.T. Aluminum induces changes in organic acids metabolism in Coffea arabica suspension cells with differential Al-tolerance. J. Inorg. Biochem. 2008, 102, 1631–1637. [Google Scholar] [CrossRef]
- Schaller, A.; Oecking, C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 1999, 11, 263–272. [Google Scholar]
- Pearce, G.; Moura, D.S.; Stratmann, J.; Ryan, C.A., Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 2001, 98, 12843–12847. [Google Scholar]
- Richards, K.D.; Schott, E.J.; Sharma, Y.K.; Davis, K.R.; Gardner, R.C. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998, 116, 409–418. [Google Scholar] [CrossRef]
- Delisle, G.; Champoux, M.; Houde, M. Characterization of oxalate oxidase and cell death in Al-sensitive and tolerant wheat roots. Plant Cell Physiol. 2001, 42, 324–333. [Google Scholar] [CrossRef]
- Soto, G.; Stritzler, M.; Lisi, C.; Alleva, K.; Pagano, M.E.; Ardila, F.; Mozzicafreddo, M.; Cuccioloni, M.; Angeletti, M.; Ayub, N.D. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J. Exp. Bot. 2011, 62, 5699–5711. [Google Scholar] [CrossRef]
- Nveawiah-Yoho, P.; Zhou, J.; Palmer, M.; Sauve, R.; Zhou, S.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Identification of Proteins for Salt Tolerance Using a Comparative Proteomics Analysis of Tomato Accessions with Contrasting Salt Tolerance. J. Am. Soc. Hortic. Sci. 2013, 138, 382–394. [Google Scholar]
- Yamamoto, Y.; Kobayashi, Y.; Devi, S.R.; Rikiishi, S.; Matsumoto, H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 2002, 128, 63–72. [Google Scholar] [CrossRef]
- Yin, L.; Mano, J.; Wang, S.; Tsuji, W.; Tanaka, K. The involvement of lipid peroxide-derived aldehydes in aluminum toxicity of tobacco roots. Plant Physiol. 2010, 152, 1406–1417. [Google Scholar] [CrossRef]
- Von Uexküll, H.R.; Mutert, E. Global extent, development and economic impact of acid soils. In Plant-Soil Interactions at Low pH: Principles and Management; Date, R.A., Grundon, N.J., Raymet, G.E., Probert, M.E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; pp. 5–19. [Google Scholar]
- Carver, B.F.; Ownby, J.D. Acid soil tolerance in wheat. Adv. Agron. 1995, 54, 117–173. [Google Scholar] [CrossRef]
- Sawyer, S. Anhydrous ammonia application and dry Soils. Available online: http://www.extension.iastate.edu/CropNews/2011/1028sawyer.htm (accessed on 1 September 2012).
| Protein accessions y | Protein name x | log2 fold w (Treated/control) | Ratio v Treated/control) |
|---|---|---|---|
| Solyc03g019820.2.1 | Aquaporin | −1.88 | 0.27 |
| Solyc06g034040.1.1 | Oleosin | −1.73 | 0.30 |
| Solyc06g072130.2.1 | Aquaporin | −1.64 | 0.32 |
| Solyc02g086490.2.1 | Oleosin | −1.61 | 0.33 |
| Solyc02g084840.2.1 | Dehydrin DHN1 | −1.53 | 0.35 |
| Solyc03g112440.1.1 | Oleosin | −1.52 | 0.35 |
| Solyc06g072670.2.1 | Short-chain dehydrogenase/reductase SDR | −1.47 | 0.36 |
| Solyc06g053740.2.1 | Ubiquitin carboxyl-terminal hydrolase | −1.42 | 0.37 |
| Solyc01g109920.2.1 | Dehydrin | −1.35 | 0.39 |
| Solyc06g065050.1.1 | Transmembrane protein 205 | −1.33 | 0.40 |
| Solyc02g077240.2.1 | Pyruvate decarboxylase | −1.30 | 0.41 |
| Solyc12g010920.1.1 | Oleosin | −1.28 | 0.41 |
| Solyc09g082330.1.1 | 7S vicilin | −1.24 | 0.42 |
| Solyc12g096930.1.1 | Caleosin | −1.18 | 0.44 |
| Solyc10g008040.2.1 | Seed biotin-containing protein SBP65 | −1.17 | 0.44 |
| Solyc11g067250.1.1 | Poly (AHRD V1 ***- B9SCR8_RICCO) | −1.11 | 0.46 |
| Solyc02g085590.2.1 | Vicilin | −1.10 | 0.47 |
| Solyc11g072380.1.1 | Vicilin-like protein | −1.09 | 0.47 |
| Solyc06g075270.2.1 | Convicilin | −1.07 | 0.48 |
| Solyc01g100390.2.1 | Pyrophosphate-energized proton pump | −1.06 | 0.48 |
| Solyc06g009210.2.1 | Ribosomal protein L19 | −1.03 | 0.49 |
| Solyc09g025210.2.1 | Legumin 11S-globulin | −1.03 | 0.49 |
| Solyc05g053140.2.1 | 26S proteasome non-ATPase regulatory subunit 13 | −1.02 | 0.49 |
| Solyc10g076510.1.1 | Pyruvate decarboxylase | −1.01 | 0.50 |
| Solyc05g053120.1.1 | Glucosyltransferase | −0.99 | 0.50 |
| Solyc08g014000.2.1 | Lipoxygenase | −0.98 | 0.51 |
| Solyc08g078850.2.1 | L-lactate dehydrogenase | −0.95 | 0.52 |
| Solyc01g009660.1.1 | Low-temperature-induced 65 kDa protein | −0.94 | 0.52 |
| Solyc01g098850.2.1 | Short-chain dehydrogenase/reductase family protein | −0.94 | 0.52 |
| Solyc06g076640.2.1 | Tubulin beta chain | −0.94 | 0.52 |
| Solyc03g116590.2.1 | Embryo-specific 3 | −0.94 | 0.52 |
| Solyc06g059740.2.1 | Alcohol dehydrogenase 2 | −0.93 | 0.53 |
| Solyc11g042800.1.1 | Late embryogenesis abundant protein | −0.93 | 0.53 |
| Solyc03g025810.2.1 | Low-temperature-induced 65 kDa protein | −0.92 | 0.53 |
| Solyc03g083970.2.1 | IQ calmodulin-binding motif family protein | −0.91 | 0.53 |
| Solyc08g013860.2.1 | NAD-dependent malic enzyme 2 | −0.90 | 0.53 |
| Solyc10g078770.1.1 | Seed maturation protein LEA 4 | −0.90 | 0.53 |
| Solyc09g090150.2.1 | Legumin 11S-globulin | −0.89 | 0.54 |
| Solyc03g112590.2.1 | Cell division protease ftsH homolog | −0.89 | 0.54 |
| Solyc04g064710.2.1 | Alcohol dehydrogenase 2 | −0.89 | 0.54 |
| Solyc00g297330.1.1 | Unknown Protein | −0.88 | 0.54 |
| Solyc07g053360.2.1 | Seed biotin-containing protein SBP65 | −0.87 | 0.55 |
| Solyc08g080480.2.1 | Unknown Protein | −0.87 | 0.55 |
| Solyc06g074750.1.1 | Histone H2B | −0.86 | 0.55 |
| Solyc12g098940.1.1 | Ubiquitin | −0.85 | 0.55 |
| Solyc07g032740.2.1 | Aspartate aminotransferase | −0.85 | 0.56 |
| Solyc01g007940.2.1 | Alanine aminotransferase 2 | −0.83 | 0.56 |
| Solyc09g065470.2.1 | Vicilin | −0.83 | 0.56 |
| Solyc09g015070.2.1 | Reductase 1 | −0.83 | 0.56 |
| Solyc12g014380.1.1 | Glucose-6-phosphate isomerase 1 | −0.81 | 0.57 |
| Solyc07g005390.2.1 | Aldehyde dehydrogenase | −0.81 | 0.57 |
| Solyc09g082340.2.1 | Vicilin-like protein | −0.80 | 0.57 |
| Solyc01g107910.2.1 | Caffeoyl CoA 3-O-methyltransferase | 0.80 | 1.74 |
| Solyc00g009020.2.1 | Mitochondrial ATP synthase | 0.80 | 1.74 |
| Solyc06g063220.2.1 | ATP synthase subunit epsilon mitochondrial | 0.80 | 1.74 |
| Solyc02g082090.2.1 | Peroxidase | 0.80 | 1.74 |
| Solyc01g102830.2.1 | Unknown Protein | 0.81 | 1.75 |
| Solyc00g147570.2.1 | Gelsolin | 0.81 | 1.75 |
| Solyc01g080510.2.1 | Os05g0406000 protein | 0.81 | 1.75 |
| Solyc08g068220.2.1 | 50S ribosomal protein L27 | 0.81 | 1.75 |
| Solyc03g078000.2.1 | High-affinity fructose transporter ght6 | 0.81 | 1.76 |
| Solyc03g096840.2.1 | Seed specific protein Bn15D1B | 0.82 | 1.76 |
| Solyc06g075810.2.1 | NADH dehydrogenase | 0.82 | 1.77 |
| Solyc06g007630.1.1 | Ferredoxin | 0.82 | 1.77 |
| Solyc05g007800.2.1 | Negatively light-regulated protein | 0.83 | 1.77 |
| Solyc11g072450.1.1 | Mitochondrial F0 ATP synthase D chain | 0.83 | 1.78 |
| Solyc10g078450.1.1 | U6 snRNA-associated Sm-like protein LSm6 | 0.83 | 1.78 |
| Solyc10g011760.2.1 | Aldose 1-epimerase family protein | 0.84 | 1.79 |
| Solyc11g066390.1.1 | Superoxide dismutase | 0.84 | 1.79 |
| Solyc03g078670.1.1 | Unknown Protein | 0.85 | 1.80 |
| Solyc05g053960.2.1 | Cysteine-rich extensin-like protein-2 | 0.85 | 1.81 |
| Solyc03g097360.2.1 | BolA-like | 0.85 | 1.81 |
| Solyc05g056020.2.1 | V-type proton ATPase subunit G 2 | 0.86 | 1.81 |
| Solyc07g063630.2.1 | Vesicle-associated membrane family protein | 0.86 | 1.82 |
| Solyc04g082590.2.1 | Canopy homolog 2 | 0.86 | 1.82 |
| Solyc02g079750.2.1 | Flavoprotein wrbA | 0.87 | 1.82 |
| Solyc02g078540.2.1 | Unknown Protein | 0.87 | 1.82 |
| Solyc11g065270.1.1 | CHCH domain containing protein | 0.87 | 1.83 |
| Solyc01g007670.2.1 | 30S ribosomal protein S7 chloroplastic | 0.87 | 1.83 |
| Solyc07g021500.1.1 | Unknown Protein | 0.87 | 1.83 |
| Solyc06g083820.2.1 | 60 ribosomal protein L14 | 0.88 | 1.84 |
| Solyc00g072400.2.1 | Peroxidase 1 | 0.88 | 1.84 |
| Solyc08g006900.2.1 | Ribosomal protein L32 | 0.88 | 1.84 |
| Solyc10g007350.2.1 | Multiprotein bridging factor 1 | 0.88 | 1.84 |
| Solyc07g055250.2.1 | Cell wall-associated hydrolase | 0.89 | 1.85 |
| Solyc08g075830.2.1 | Peroxidase 27 | 0.89 | 1.85 |
| Solyc05g041610.1.1 | Caffeoyl-CoA O-methyltransferase | 0.89 | 1.86 |
| Solyc07g008350.2.1 | Porin/voltage-dependent anion-selective channel protein | 0.89 | 1.86 |
| Solyc11g011340.1.1 | Alcohol dehydrogenase | 0.90 | 1.86 |
| Solyc12g094700.1.1 | Cathepsin B-like cysteine proteinase | 0.90 | 1.87 |
| Solyc01g049960.2.1 | Unknown Protein | 0.92 | 1.89 |
| Solyc03g114970.2.1 | Nitrilase associated protein-like | 0.92 | 1.89 |
| Solyc09g082710.2.1 | Histone H2A | 0.92 | 1.89 |
| Solyc08g016420.2.1 | Prefoldin subunit 6 | 0.92 | 1.90 |
| Solyc03g025850.2.1 | Remorin 1 | 0.93 | 1.91 |
| Solyc01g103220.2.1 | Cytochrome c | 0.94 | 1.92 |
| Solyc07g065640.2.1 | RPM1 interacting protein 4 transcript 2 | 0.94 | 1.92 |
| Solyc05g056290.2.1 | Acetyl-CoA carboxylase biotin carboxyl carrier protein | 0.95 | 1.93 |
| Solyc04g049330.2.1 | V-type proton ATPase subunit G 1 | 0.96 | 1.94 |
| Solyc01g091130.2.1 | Nitroreductase | 0.96 | 1.95 |
| Solyc01g095150.2.1 | Late embryogenesis abundant protein | 0.97 | 1.96 |
| Solyc07g005240.2.1 | FAD-dependent oxidoreductase family protein | 0.97 | 1.97 |
| Solyc01g090360.2.1 | Non-specific lipid-transfer protein | 0.98 | 1.97 |
| Solyc01g095050.2.1 | Negatively light-regulated protein | 0.98 | 1.98 |
| Solyc04g082010.1.1 | Plastocyanin | 1.00 | 1.99 |
| Solyc11g008990.1.1 | Phage shock protein A PspA | 1.00 | 2.00 |
| Solyc04g007750.2.1 | Major latex-like protein | 1.01 | 2.01 |
| Solyc03g113730.2.1 | B12D protein | 1.03 | 2.04 |
| Solyc08g013930.2.1 | Peroxidase family protein | 1.04 | 2.05 |
| Solyc01g088140.2.1 | Unknown Protein | 1.04 | 2.05 |
| Solyc02g085230.2.1 | Nucleolar protein 6 | 1.04 | 2.06 |
| Solyc06g036380.1.1 | Ulp1 protease family C-terminal catalytic domain containing protein | 1.04 | 2.06 |
| Solyc12g019040.1.1 | Exostosin family protein | 1.04 | 2.06 |
| Solyc03g116060.2.1 | Gibberellin-regulated protein | 1.04 | 2.06 |
| Solyc06g054520.1.1 | 3-hydroxyisobutyryl-CoA hydrolase | 1.05 | 2.07 |
| Solyc02g043900.1.1 | Unknown Protein | 1.06 | 2.08 |
| Solyc07g041490.1.1 | Stress responsive alpha-beta barrel domain protein | 1.07 | 2.09 |
| Solyc04g071580.2.1 | Unknown Protein | 1.08 | 2.12 |
| Solyc08g008330.2.1 | Unknown Protein | 1.09 | 2.13 |
| Solyc09g074890.1.1 | Rapid alkalinization factor 1 | 1.11 | 2.15 |
| Solyc04g024840.2.1 | GDSL esterase/lipase 1 | 1.11 | 2.16 |
| Solyc04g074900.2.1 | 40S ribosomal protein S21 | 1.12 | 2.17 |
| Solyc06g054250.2.1 | 5&apos-nucleotidase surE | 1.14 | 2.21 |
| Solyc02g092270.2.1 | NADH dehydrogenase | 1.14 | 2.21 |
| Solyc12g008950.1.1 | At1g17490/F1L3_4 | 1.15 | 2.22 |
| Solyc10g076240.1.1 | Peroxidase 1 | 1.21 | 2.31 |
| Solyc03g113580.1.1 | Germin-like protein | 1.26 | 2.40 |
| Solyc03g118110.2.1 | Succinate dehydrogenase assembly factor 2, mitochondrial | 1.30 | 2.45 |
| Solyc07g054960.1.1 | Myb-related transcription factor | 1.35 | 2.55 |
| Solyc10g005660.2.1 | COP9 signalosome subunit 6 | 1.35 | 2.55 |
| Solyc11g010160.1.1 | Cc-nbs-lrr, resistance protein | 1.35 | 2.55 |
| Solyc06g062770.2.1 | At1g17490/F1L3_4 | 1.36 | 2.56 |
| Solyc03g117810.2.1 | Phosphate import ATP-binding protein pstB 1 | 1.36 | 2.57 |
| Solyc11g066270.1.1 | Xyloglucan endotransglucosylase/hydrolase 9 | 1.37 | 2.59 |
| Solyc05g007090.2.1 | Zinc knuckle | 1.37 | 2.59 |
| Solyc01g107990.2.1 | MAP protein kinase-like protein | 1.42 | 2.68 |
| Solyc00g015000.1.1 | DNA (Cytosine-5-)-methyltransferase 3 | 1.55 | 2.93 |
| Solyc02g093230.2.1 | Caffeoyl-CoA O-methyltransferase | 1.55 | 2.93 |
| Solyc04g028490.1.1 | Ulp1 protease family C-terminal catalytic domain containing protein | 1.55 | 2.93 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Okekeogbu, I.; Ye, Z.; Sangireddy, S.R.; Li, H.; Bhatti, S.; Hui, D.; Zhou, S.; Howe, K.J.; Fish, T.; Yang, Y.; et al. Effect of Aluminum Treatment on Proteomes of Radicles of Seeds Derived from Al-Treated Tomato Plants. Proteomes 2014, 2, 169-190. https://doi.org/10.3390/proteomes2020169
Okekeogbu I, Ye Z, Sangireddy SR, Li H, Bhatti S, Hui D, Zhou S, Howe KJ, Fish T, Yang Y, et al. Effect of Aluminum Treatment on Proteomes of Radicles of Seeds Derived from Al-Treated Tomato Plants. Proteomes. 2014; 2(2):169-190. https://doi.org/10.3390/proteomes2020169
Chicago/Turabian StyleOkekeogbu, Ikenna, Zhujia Ye, Sasikiran Reddy Sangireddy, Hui Li, Sarabjit Bhatti, Dafeng Hui, Suping Zhou, Kevin J. Howe, Tara Fish, Yong Yang, and et al. 2014. "Effect of Aluminum Treatment on Proteomes of Radicles of Seeds Derived from Al-Treated Tomato Plants" Proteomes 2, no. 2: 169-190. https://doi.org/10.3390/proteomes2020169





