Protein Profiling Reveals Novel Proteins in Pollen and Pistil of W22 (ga1; Ga1) in Maize
Abstract
:1. Introduction
2. Experimental
2.1. Plant Materials and Genetic Background
2.2. Protein Extraction and Electrophoresis
2.3. In-gel Digestion
2.4. MS/MS Analysis and Bioinformatics
3. Results and Discussion
3.1. Protein Expression on SDS-PAGE
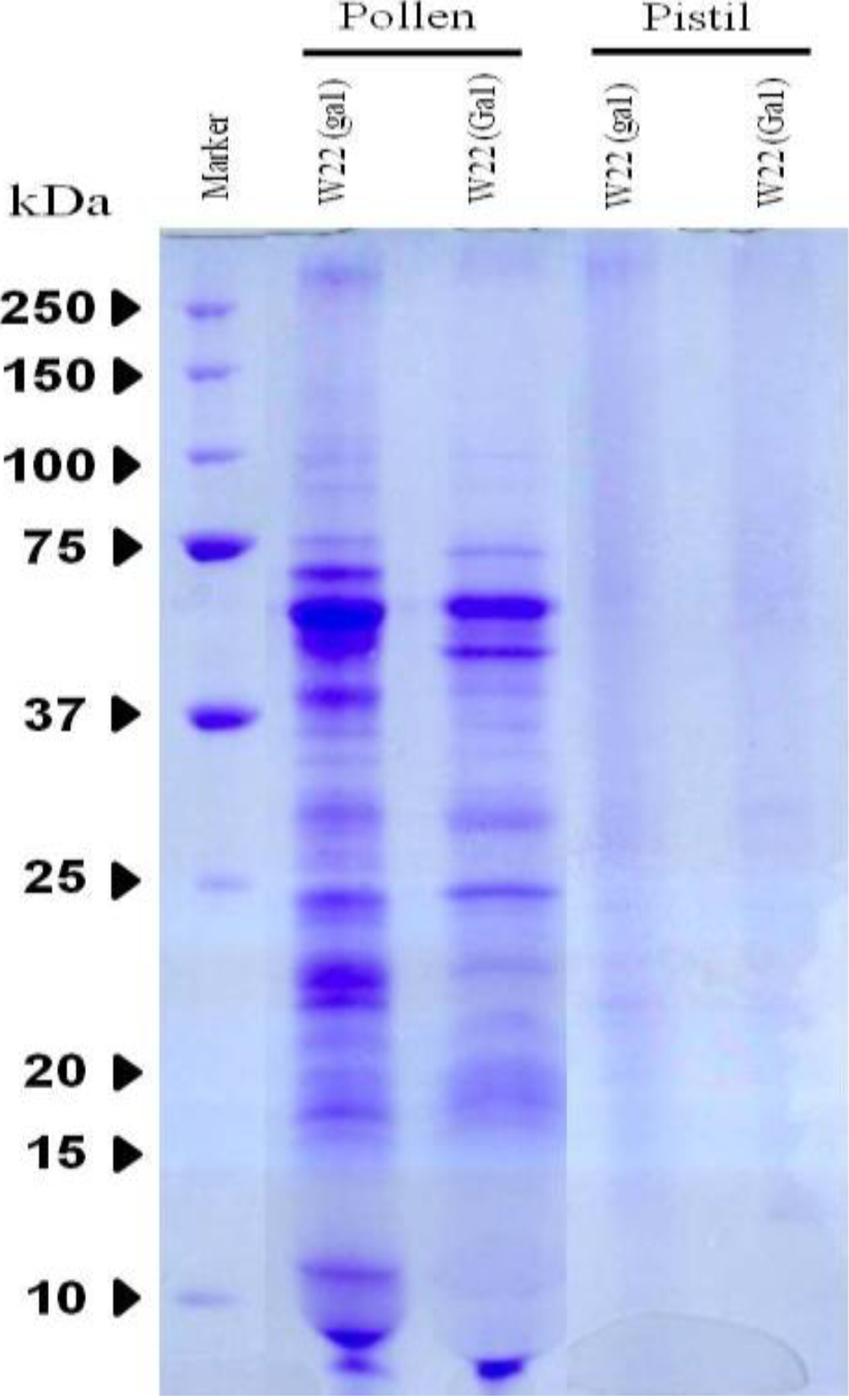
3.2. Specific Protein Analysis of Identifying Proteins from Pollen
3.3. Specific Protein Analysis of Identified Proteins from Pistil
| AN 1 | Protein Description | W22 (ga1) | W22 (Ga1) | PS 2 | MW.3 | PM 4 | pI.5 | PC.6 | MS-MS Ion Score | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pollen | Pistil | Pollen | Pistil | ||||||||
| Hydrolase activity | |||||||||||
| P26216 | Exopolygalacturonase | √ | × | √ | × | 79 | 43,416 | 6.95 | 4 | 11.7 | 64.93 |
| P35339 | Exopolygalacturonase | √ | × | √ | × | 63 | 43,269 | 8.44 | 5 | 17.8 | 67.85 |
| P35338 | Exopolygalacturonase | √ | × | √ | × | 79 | 43,387 | 6.59 | 5 | 15.1 | 66.03 |
| Q41803 | Elongation factor 1-alpha | √ | × | √ | × | 96 | 49,202 | 9.19 | 7 | 23 | 46.8 |
| P29022 | Endochitinase A | √ | × | √ | × | 103 | 29,106 | 8.3 | 3 | 22.5 | 69.56 |
| P29023 | Endochitinase B (Fragment) | √ | × | √ | × | 61 | 28,147 | 8.94 | 2 | 14.9 | 59.82 |
| P10593 | Albumin b-32 | √ | × | × | × | 36 | 32,408 | 5.38 | 3 | 20.8 | 46.86 |
| P25891 | Ribosome-inactivating protein 3 | √ | × | √ | × | 30 | 33,236 | 5.83 | 3 | 15.3 | 36.78 |
| Nucleotide binding | |||||||||||
| Q43298 | Chaperonin CPN60-2, mitochondrial | √ | × | × | × | 31 | 60,897 | 5.67 | 2 | 6.2 | 9.26 |
| P49094 | Asparagine synthetase | × | √ | × | × | 21 | 66,535 | 5.83 | 2 | 6.1 | 21.43 |
| P05494 | ATP synthase subunit alpha, mitochondrial | × | × | √ | × | 102 | 55,146 | 8 | 5.85 | 19.3 | 34.9 |
| P49106 | 14-3-3-like protein GF14-6 | × | × | × | √ | 157 | 29,644 | 3 | 4.76 | 17.6 | 48.74 |
| O24594 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | × | × | × | √ | 28 | 60,892 | 2 | 6.77 | 6 | 3.07 |
| Nucleic acid binding | |||||||||||
| P30755 | Histone H2B.1 | × | √ | × | × | 32 | 16,410 | 10 | 5 | 39.1 | 16.68 |
| P40280 | Histone H2A | × | √ | √ | × | 65 | 16,417 | 10.59 | 2 | 28.3 | 70.53 |
| Q8S4P5 | Histone-lysine N-methyltransferase EZ2 | × | √ | × | √ | 24 | 99,916 | 8.47 | 7 | 11 | 9.82 |
| Q8S4P6 | Histone-lysine N-methyltransferase EZ1 | × | √ | × | × | 14 | 103,703 | 8.85 | 3 | 4.7 | 2.64 |
| P49120 | Histone H2B.4 | × | × | √ | × | 65 | 15,173 | 4 | 10.02 | 31.4 | 4.97 |
| Catalytic activity | |||||||||||
| P80608 | Cysteine synthase | × | × | √ | × | 31 | 34,185 | 2 | 5.91 | 10.5 | 5.12 |
| P23225 | Ferredoxin-dependent glutamate synthase, chloroplastic | × | × | √ | × | 18 | 175,063 | 5 | 6.21 | 5.8 | 8.66 |
| Q41769 | Acetolactate synthase 2 | × | √ | × | × | 31 | 68,982 | 6.48 | 3 | 9.1 | 29.41 |
| P30792 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase | × | √ | × | √ | 59 | 60,582 | 5.29 | 3 | 9.7 | 67.15 |
| Antioxidant activity | |||||||||||
| P18122 | Catalase isozyme 1 | × | √ | × | × | 27 | 56,841 | 7.4 | 2 | 9.8 | 26.82 |
| P18123 | Catalase isozyme 3 | × | √ | × | × | 27 | 56,760 | 6.47 | 2 | 4.8 | 5.02 |
| A2SZW8 | 1-Cys peroxiredoxin PER1 | × | × | √ | × | 28 | 24,890 | 2 | 6.31 | 10.5 | 38.51 |
| Hydrolase activity | |||||||||||
| P04709 | ADP, ATP carrier protein 1 | × | √ | × | × | 41 | 42,365 | 9.85 | 4 | 11.1 | 22.01 |
| 857 | ADP, ATP carrier protein 2 | × | × | √ | × | 56 | 42,306 | 4 | 9.85 | 12.4 | 4.23 |
| Protein binding | |||||||||||
| P35083 | Profilin-3 | √ | × | × | × | 60 | 14,228 | 4.91 | 2 | 29 | 60.41 |
| Q01526 | 14-3-3-like protein GF14-12 | × | √ | × | × | 149 | 29,618 | 4.75 | 3 | 18 | 8.99 |
| Oxidoreductase activity | |||||||||||
| P09315 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic | × | × | × | √ | 31 | 42,840 | 2 | 7 | 8.7 | 30.94 |
| P08735 | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic 1 | × | × | × | √ | 44 | 36,500 | 3 | 6.46 | 13.9 | 5.2 |
| Transporter activity | |||||||||||
| P19023 | ATP synthase subunit beta, mitochondrial | × | × | √ | × | 128 | 59,067 | 8 | 6.01 | 25.9 | 25.94 |
| P28523 | Casein kinase II subunit alpha | × | × | × | √ | 16 | 39,205 | 3 | 8.41 | 14.8 | 16.2 |
| Isomerase activity | |||||||||||
| P21569 | Peptidyl-prolyl cis-trans isomerase | × | × | √ | × | 127 | 18,337 | 2 | 8.91 | 18 | 36.38 |
| P49105 | Glucose-6-phosphate isomerase | × | √ | √ | × | 36 | 62,198 | 2 | 6.96 | 4.1 | 1.62 |
| Enzyme regulator activity | |||||||||||
| P13867 | Alpha-amylase/trypsin inhibitor | × | √ | × | √ | 64 | 22,060 | 8.16 | 2 | 15 | 62.82 |
| Signal transducer activity | |||||||||||
| Q9C9W9 | Adagio protein 3 | × | × | √ | × | 23 | 69,019 | 3 | 6.06 | 2.3 | 9.6 |
| Unknown | |||||||||||
| P0C1Y5 | Expansin-B11 | √ | × | √ | × | 174 | 28,943 | 8.44 | 4 | 13.8 | 39.64 |
| Q8VZY6 | Polycomb group protein FIE2 | × | √ | × | × | 27 | 42,475 | 5.89 | 2 | 15.3 | 9.65 |
| Q07154 | Expansin-B9 | × | × | √ | × | 200 | 29,062 | 5 | 9.01 | 21.6 | 39.5 |
| P58738 | Expansin-B1 | × | × | √ | × | 31 | 29,066 | 4 | 8.99 | 18.6 | 39.5 |
| P46517 | Late embryogenesis abundant protein EMB564 | × | × | × | √ | 55 | 9678 | 2 | 6.6 | 27.5 | 55.46 |
| B6TYV8 | Cell number regulator 2 | × | × | × | √ | 18 | 19,222 | 2 | 7.37 | 17.7 | 18.17 |
3.4. Cross-Correlation and Functional Distribution of Identified Proteins from Pollen and Pistil

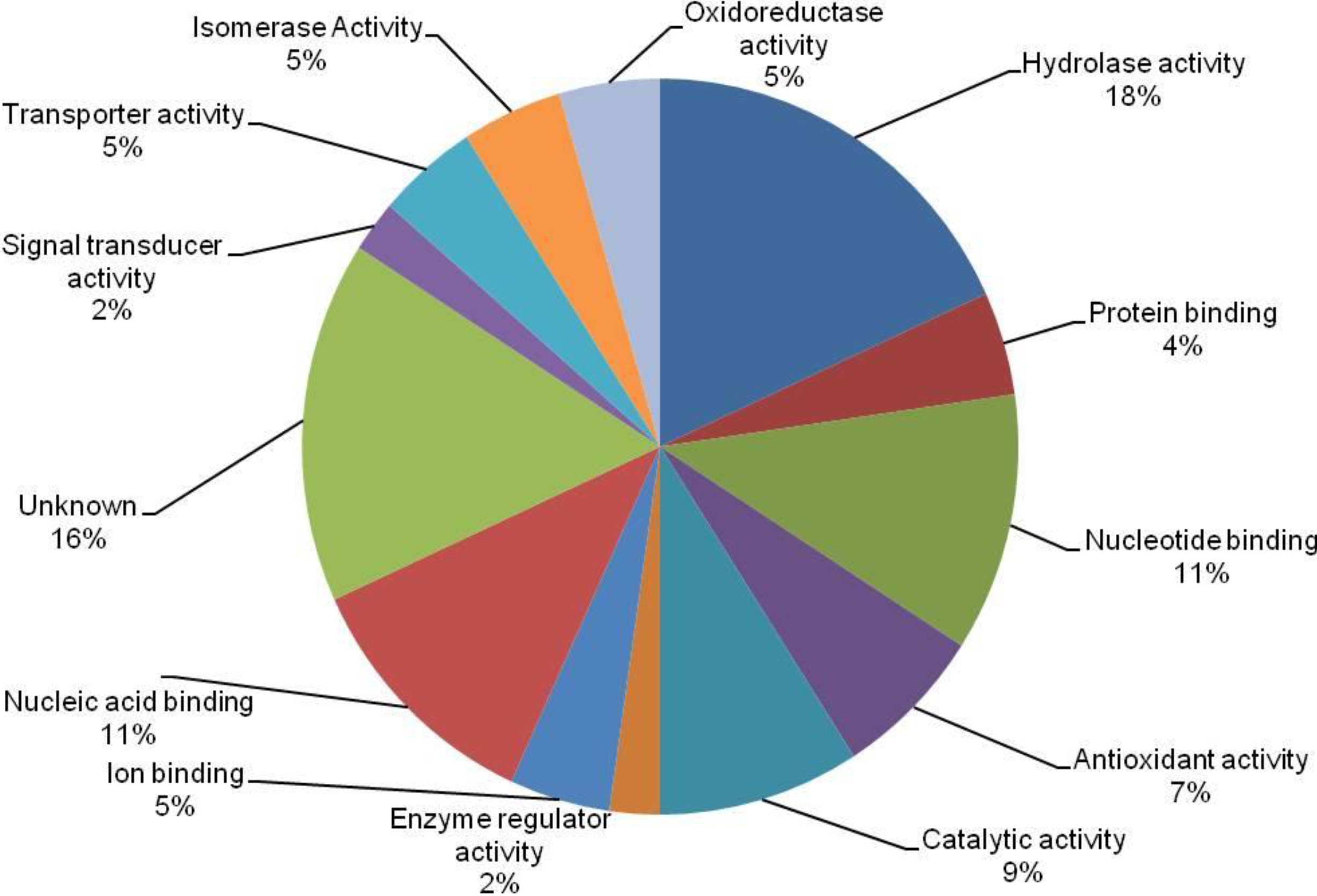
3.5. The Implication of Differentially Expressed Proteins from Pollen and Pistil of Maize
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cheung, A.Y. Pollen-pistil interactions in compatible pollination. Proc. Natl. Acad. Sci. USA 1995, 92, 3077–3080. [Google Scholar] [CrossRef]
- Kermicle, J.L.; Evans, M.M.S. The Zea mays sexual compatibility gene ga2: Naturally occurring alleles, their distribution, and role in reproductive isolation. J. Hered. 2010, 101, 737–749. [Google Scholar] [CrossRef]
- Mangelsdorf, P.C.; Jones, D.F. The expression of mendelian factors in the gametophyte of maize. Genetics 1926, 11, 423–455. [Google Scholar]
- Burnham, C.R. Cytogenetic studies of a case of pollen abortion in maize. Genetics 1941, 26, 460–468. [Google Scholar]
- Cameron, D.R.; Moav, R.M. Inheritance in Nicotiana tabacum XXVII. Pollen killer, an alien genetic locus inducing abortion of microspores not carrying it. Genetics 1957, 42, 326–335. [Google Scholar]
- Loegering, W.Q.; Sears, E.R. Distorted inheritance of stem-rust resistance of timstein wheat caused by a pollen-killing gene. Can. J. Genet. Cytol. 1963, 5, 65–72. [Google Scholar]
- Rick, C.M. Abortion of male and female gametes in the tomato determined by allelic interaction. Genetics 1966, 53, 85–96. [Google Scholar]
- Allard, R.W. An additional gametophyte factor in the lima bean. Der Zücht. 1963, 33, 212–216. [Google Scholar]
- Ramage, R.T. Heterosis and hybrid seed production in barley. In Heterosis; Frankel, R., Ed.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 1983; Volume 6, pp. 71–93. [Google Scholar]
- Dresselhaus, T.; Lausser, A.; Márton, M.L. Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann. Bot. 2011, 108, 727–737. [Google Scholar] [CrossRef]
- Nelson, O. The gametophyte factors of maize. In The Maize Handbook; Freeling, M., Walbot, V., Eds.; Springer New York: New York, NY, USA, 1994; pp. 496–503. [Google Scholar]
- Li, X.M.; Sang, Y.L.; Zhao, X.Y.; Zhang, X.S. High-throughput sequencing of small RNAs from pollen and silk and characterization of miRNAs as candidate factors involved in pollen-silk interactions in maize. PLoS One 2013, 8, e72852. [Google Scholar]
- Chen, Y.; Chen, T.; Shen, S.; Zheng, M.; Guo, Y.; Lin, J.; Baluška, F.; Šamaj, J. Differential display proteomic analysis of Picea meyeri pollen germination and pollen-tube growth after inhibition of actin polymerization by latrunculin B. Plant J. 2006, 47, 174–195. [Google Scholar] [CrossRef]
- Fernando, D.D. Characterization of pollen tube development in Pinus strobus (eastern white pine) through proteomic analysis of differentially expressed proteins. Proteomics 2005, 5, 4917–4926. [Google Scholar] [CrossRef]
- Holmes-Davis, R.; Tanaka, C.K.; Vensel, W.H.; Hurkman, W.J.; McCormick, S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 2005, 5, 4864–4884. [Google Scholar] [CrossRef]
- Imin, N.; Kerim, T.; Weinman, J.J.; Rolfe, B.G. Characterisation of rice anther proteins expressed at the young microspore stage. Proteomics 2001, 1, 1149–1161. [Google Scholar] [CrossRef]
- Imin, N.; Kerim, T.; Weinman, J.J.; Rolfe, B.G. Low temperature treatment at the young microspore stage induces protein changes in rice anthers. Mol. Cell. Proteomics 2006, 5, 274–292. [Google Scholar]
- Noir, S.; Bräutigam, A.; Colby, T.; Schmidt, J.; Panstruga, R. A reference map of the Arabidopsis thaliana mature pollen proteins. Biochem. Biophys. Res. Commun. 2005, 337, 1257–1266. [Google Scholar] [CrossRef]
- Kamal, A.H.M.; Jang, I.D.; Kim, D.E.; Suzuki, T.; Chung, K.Y.; Choi, J.S.; Lee, M.S.; Park, C.H.; Park, S.U.; Lee, S.H.; et al. Proteomics analysis of embryo and endosperm from mature common buckwheat seeds. J. Plant Biol. 2011, 54, 81–91. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
- Fonzo, N.; Hartings, H.; Brembilla, M.; Motto, M.; Soave, C.; Navarro, E.; Palau, J.; Rhode, W.; Salamini, F. The b-32 protein from maize endosperm, an albumin regulated by the o2 locus: Nucleic acid (cDNA) and amino acid sequences. Mol. Gen. Genet. MCG 1988, 212, 481–487. [Google Scholar] [CrossRef]
- Antes, I.; Chandler, D.; Wang, H.; Oster, G. The unbinding of ATP from F1-ATPase. Biophys. J. 2003, 85, 695–706. [Google Scholar] [CrossRef]
- Lee, K.O.; Jang, H.H.; Jung, B.G.; Chi, Y.H.; Lee, J.Y.; Choi, Y.O.; Lee, J.R.; Lim, C.O.; Cho, M.J.; Lee, S.Y. Rice 1cys-peroxiredoxin over-expressed in transgenic tobacco does not maintain dormancy but enhances antioxidant activity. FEBS Lett. 2000, 486, 103–106. [Google Scholar] [CrossRef]
- Hoang, T.K.H.; Akihiro, N. Mitochondrial proteomic analysis of cam plants, Ananas comosus and Kalanchoe pinnata. Ann. Biol. Res. 2012, 3, 88–97. [Google Scholar]
- Takahashi, H.; Saito, K. Subcellular localization of spinach cysteine synthase isoforms and regulation of their gene expression by nitrogen and sulfur. Plant Physiol. 1996, 112, 273–280. [Google Scholar]
- Mendoza-Cózatl, D.; Loza-Tavera, H.; Hernández-Navarro, A.; Moreno-Sánchez, R. Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol. Rev. 2005, 29, 653–671. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Zybailov, B.; Friso, G.; Kim, J.; Rudella, A.; Rodríguez, V.R.; Asakura, Y.; Sun, Q.; van Wijk, K.J. Large scale comparative proteomics of a chloroplast Clp protease mutant reveals folding stress, altered protein homeostasis, and feedback regulation of metabolism. Mol. Cell. Proteomics 2009, 8, 1789–1810. [Google Scholar] [CrossRef]
- Mittler, R.; Zilinskas, B.A. Regulation of pea cytosolic ascorbate peroxidase and other antioxidant enzymes during the progression of drought stress and following recovery from drought. Plant J. 1994, 5, 397–405. [Google Scholar]
- Jiang, S.-S.; Liang, X.-N.; Li, X.; Wang, S.-L.; Lv, D.-W.; Ma, C.-Y.; Li, X.-H.; Ma, W.-J.; Yan, Y.-M. Wheat drought-responsive grain proteome analysis by linear and nonlinear 2-DE and MALDI-TOF mass spectrometry. Int. J. Mol. Sci. 2012, 13, 16065–16083. [Google Scholar] [CrossRef]
- Chipman, D.; Barak, Z.E.; Schloss, J.V. Biosynthesis of 2-aceto-2-hydroxy acids: Acetolactate synthases and acetohydroxyacid synthases. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol 1998, 1385, 401–419. [Google Scholar] [CrossRef]
- Dailey, F.E.; Cronan, J.E. Acetohydroxy acid synthase I, a required enzyme for isoleucine and valine biosynthesis in Escherichia coli K-12 during growth on acetate as the sole carbon source. J. Bacteriol. 1986, 165, 453–460. [Google Scholar]
- Abbatiello, S.E.; Pan, Y.-X.; Zhou, M.; Wayne, A.S.; Veenstra, T.D.; Hunger, S.P.; Kilberg, M.S.; Eyler, J.R.; Richards, N.G.J.; Conrads, T.P. Mass spectrometric quantification of asparagine synthetase in circulating leukemia cells from acute lymphoblastic leukemia patients. J. Proteomics 2008, 71, 61–70. [Google Scholar] [CrossRef]
- Ichimura, T.; Isobe, T.; Okuyama, T.; Takahashi, N.; Araki, K.; Kuwano, R.; Takahashi, Y. Molecular cloning of cdna coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc. Natl. Acad. Sci. USA 1988, 85, 7084–7088. [Google Scholar] [CrossRef]
- Sobhanian, H.; Razavizadeh, R.; Nanjo, Y.; Ehsanpour, A.A.; Jazii, F.R.; Motamed, N.; Komatsu, S. Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci. 2010, 8, 1477–5956. [Google Scholar]
- Holmberg, N.; Bülow, L. Improving stress tolerance in plants by gene transfer. Trends Plant Sci. 1998, 3, 61–66. [Google Scholar] [CrossRef]
- Amara, I.; Odena, A.; Oliveira, E.; Moreno, A.; Masmoudi, K.; Pagès, M.; Goday, A. Insights into maize lea proteins: From proteomics to functional approaches. Plant Cell Physiol. 2012, 53, 312–329. [Google Scholar] [CrossRef]
- Jockusch, B.M.; Murk, K.; Rothkegel, M. The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 2007, 159, 131–149. [Google Scholar]
- Dong, J.; Radau, B.; Otto, A.; Müller, E.-C.; Lindschau, C.; Westermann, P. Profilin I attached to the golgi is required for the formation of constitutive transport vesicles at the trans-golgi network. Biochim. Biophys. Acta (BBA) Mol. Cell. Res. 2000, 1497, 253–260. [Google Scholar] [CrossRef]
- Rothkegel, M.; Mayboroda, O.; Rohde, M.; Wucherpfennig, C.; Valenta, R.; Jockusch, B.M. Plant and animal profilins are functionally equivalent and stabilize microfilaments in living animal cells. J. Cell Sci. 1996, 109, 83–90. [Google Scholar]
- Jaquinod, M.; Villiers, F.; Kieffer-Jaquinod, S.; Hugouvieux, V.; Bruley, C.; Garin, J.; Bourguignon, J. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 2007, 6, 394–412. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yu, J.; Roy, S.K.; Kamal, A.H.M.; Cho, K.; Kwon, S.-J.; Cho, S.-W.; So, Y.-S.; Holland, J.B.; Woo, S.H. Protein Profiling Reveals Novel Proteins in Pollen and Pistil of W22 (ga1; Ga1) in Maize. Proteomes 2014, 2, 258-271. https://doi.org/10.3390/proteomes2020258
Yu J, Roy SK, Kamal AHM, Cho K, Kwon S-J, Cho S-W, So Y-S, Holland JB, Woo SH. Protein Profiling Reveals Novel Proteins in Pollen and Pistil of W22 (ga1; Ga1) in Maize. Proteomes. 2014; 2(2):258-271. https://doi.org/10.3390/proteomes2020258
Chicago/Turabian StyleYu, Jin, Swapan Kumar Roy, Abu Hena Mostafa Kamal, Kun Cho, Soo-Jeong Kwon, Seong-Woo Cho, Yoon-Sup So, James B. Holland, and Sun Hee Woo. 2014. "Protein Profiling Reveals Novel Proteins in Pollen and Pistil of W22 (ga1; Ga1) in Maize" Proteomes 2, no. 2: 258-271. https://doi.org/10.3390/proteomes2020258







