Extraction and Characterization of Extracellular Proteins and Their Post-Translational Modifications from Arabidopsis thaliana Suspension Cell Cultures and Seedlings: A Critical Review
Abstract
:1. Introduction
- (1)
- The cell wall, having no discrete membrane, is not a bounded organelle [1,4]. Instead, the cell wall may be viewed as a continuum, which makes it difficult to isolate only the cell wall and apoplastic fluid proteome without contamination by intracellular proteins. During extraction, many intracellular proteins can bind to the charged polysaccharides of the cell wall matrix and be mistakenly included in the cell wall fraction [4,27]. In addition, some bona fide apoplastic proteins may not associate directly with the cell wall, and therefore be discarded as part of the intracellular fraction [4].
- (2)
- As CWPs only comprise 5%–10% of the cell wall mass, challenges arise in enriching the CWPs, particularly since they can be embedded within the polysaccharide matrix. Some CWPs interact with the cell wall via non-covalent linkages, such as ionic, hydrophobic, or van der Waals interactions, while other CWPs can be covalently cross-linked with the polysaccharides, thus forming insoluble networks [1,4,5,28,29,30]. To date, no single protocol has successfully extracted all CWPs due to the high degree of heterogeneity by which they can associate with other cell wall components [27]. In addition, the plant cell wall varies both spatially and temporally, further confounding the elucidation of a complete cell wall proteome [1,4].
- (3)
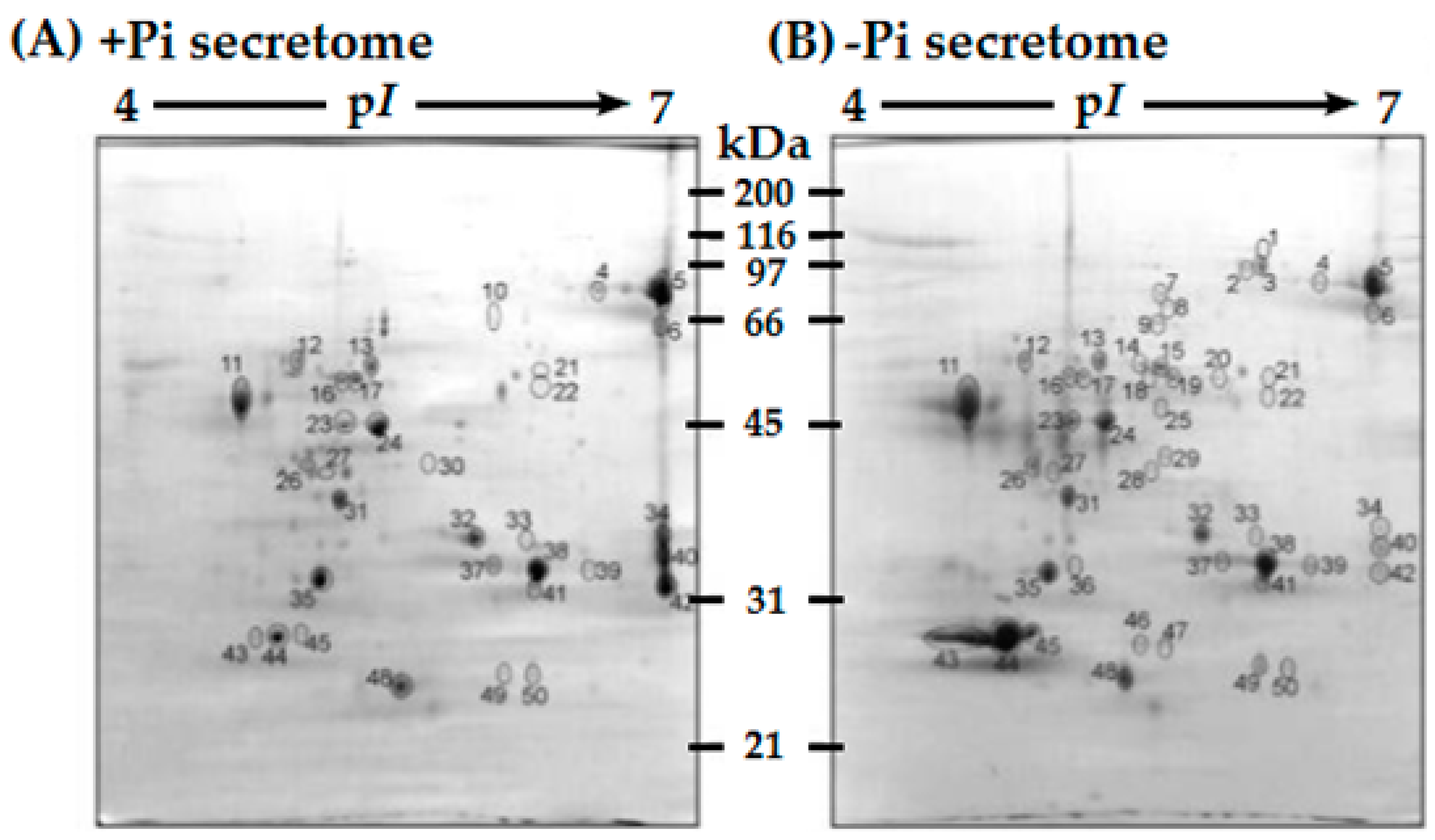
- Once CWPs have been successfully extracted, it can be difficult to separate and identify them. Most CWPs are basic, making the conventional two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) method of protein separation less effective [1]. Furthermore, low-abundance proteins or small peptide hormones are generally not detected using this traditional method of protein separation; as such, many important CWPs may be overlooked or missed completely. Additionally, PTMs of CWPs can complicate both protein separation, and protein identification and characterization using bioinformatic tools [1,3,5,6,22,23,24,26].
- (4)
- Finally, studies aiming to identify the extracellular proteome are often plagued by so-called non-canonical CWPs [1,31,32]. Despite evidence of minimal intracellular contamination, these proteins lacking the classical signal peptide that would target them to the secretory pathway persistently appear in CWP extracts, with an estimated 40%–70% of proteins identified in secretome studies falling within this non-canonical grouping [32]. Some of these proteins may indeed be intracellular contaminants; however, recent evidence suggest that a plant-specific exocyst-positive organelle may be responsible for the unconventional secretion of certain CWPs [5,32,33]. Leaderless secretion of some proteins also occurs in both mammals and yeast, and may be common to all eukaryotes [31].
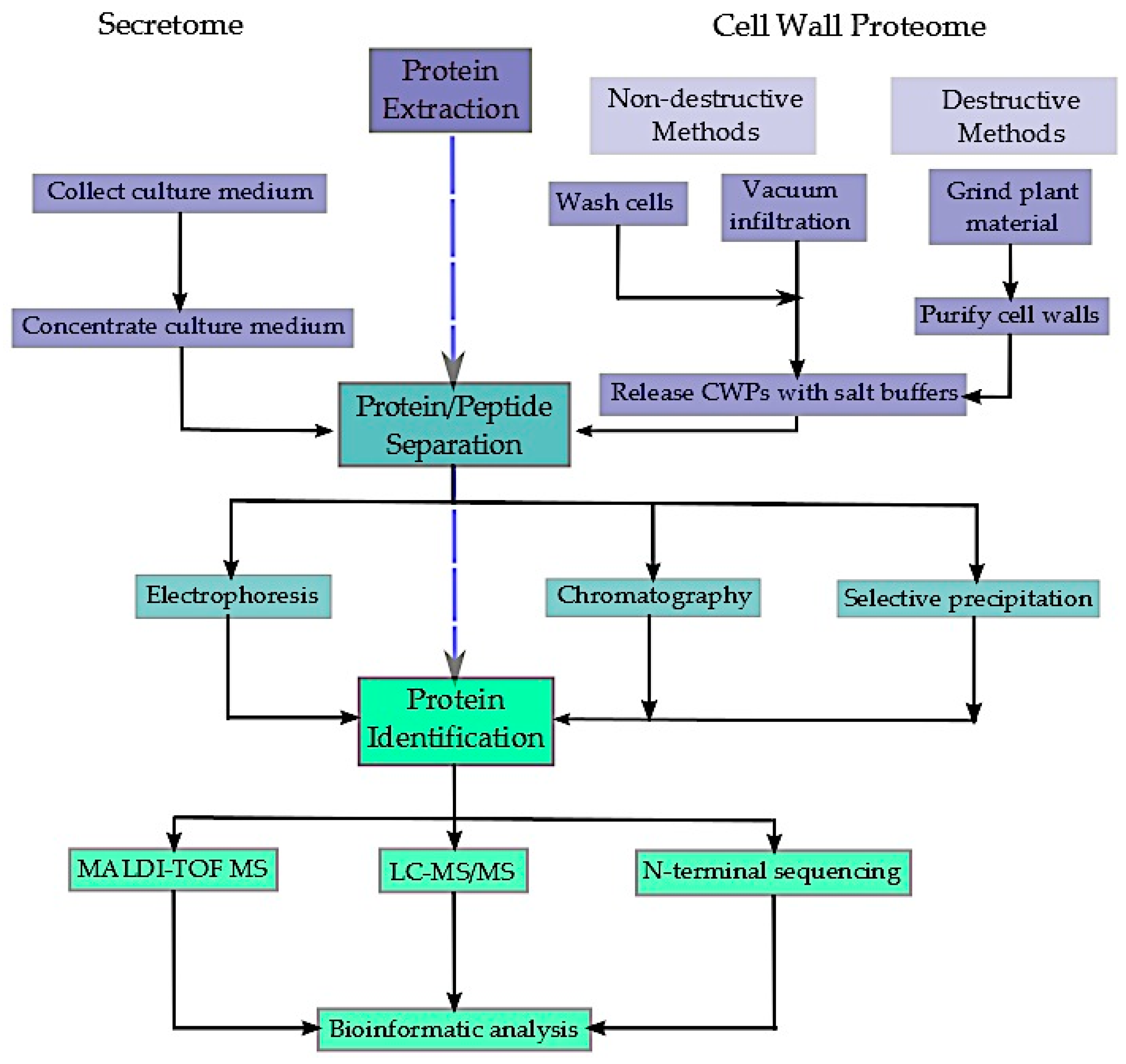
2. Extraction and Enrichment of the Extracellular Proteome
2.1. The Secretome
2.2. The Cell Wall Proteome
3. Overcoming the Challenges of Identifying the Extracellular Proteome
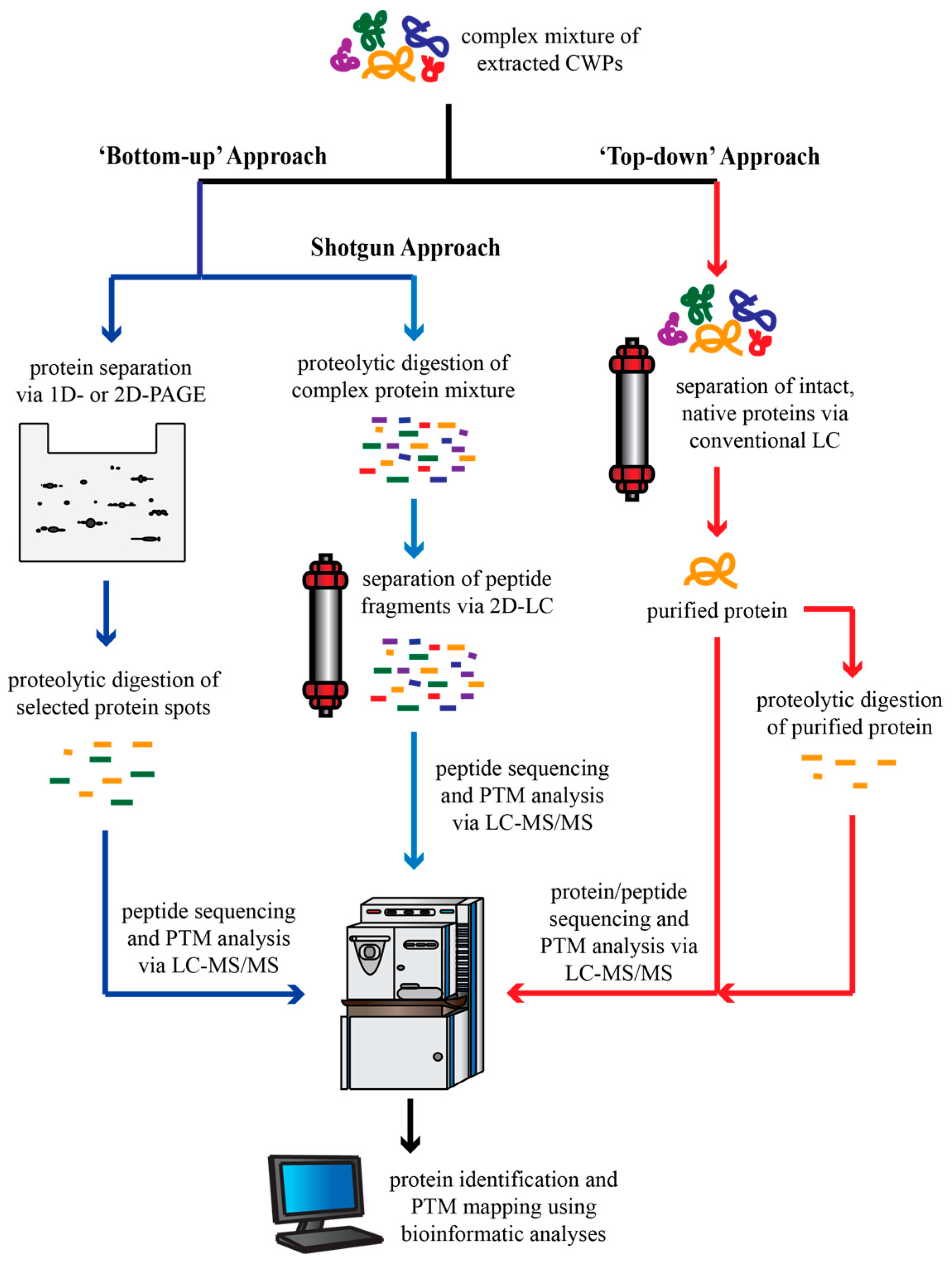
3.1. Advances in Protein Separation Techniques
3.2. The Advantages of Targeted Proteomics
3.3. Confirming Protein Localization
4. Post-translational Modifications of Cell Wall Proteins
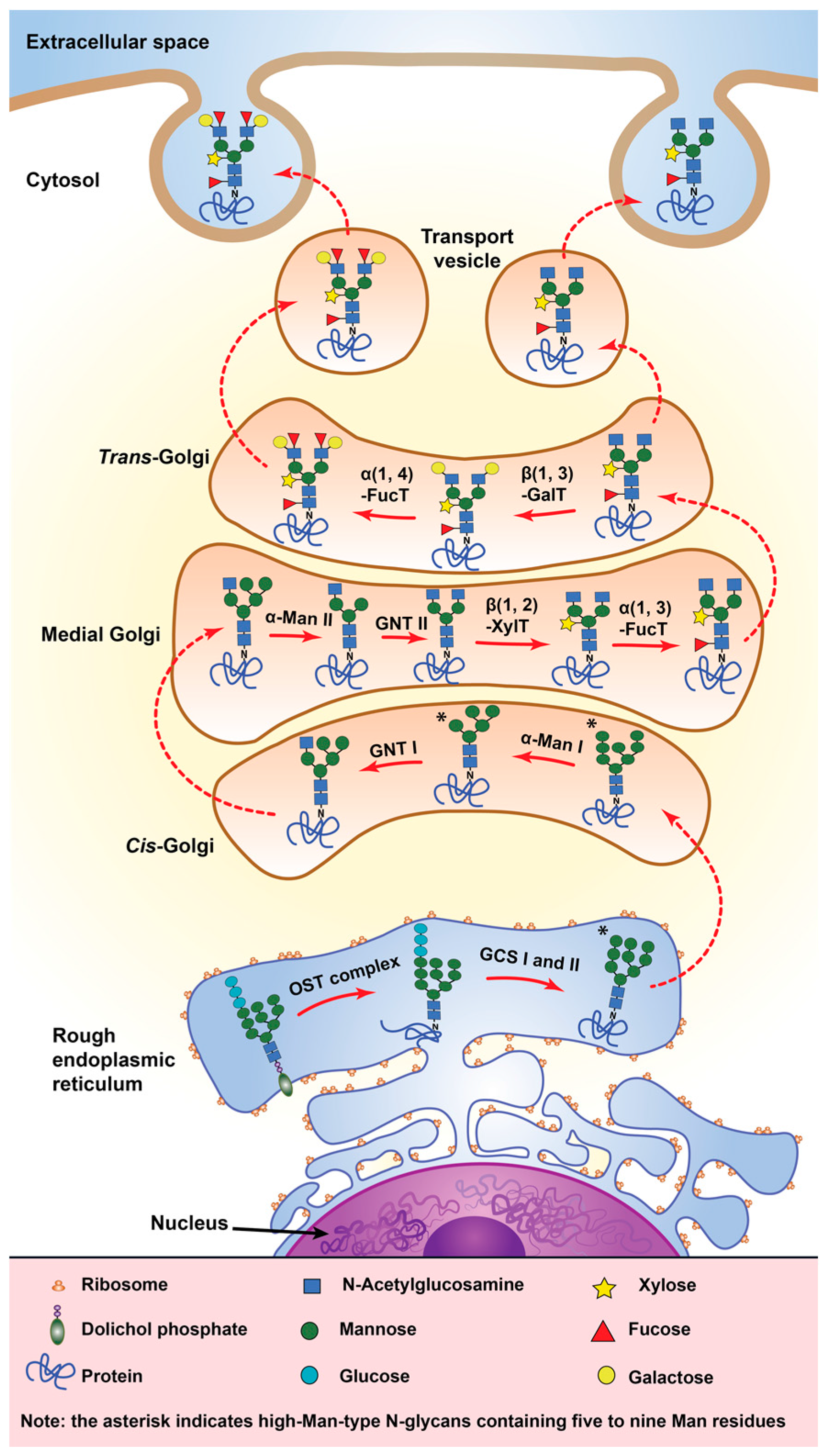
4.1. Glycosylation
4.2. Phosphorylation
- Where is/are the site(s) of phosphorylation? Mapping the specific amino-acid residue of a protein that has been phosphorylated in vivo provides fundamental information regarding the mechanistic details, as residues flanking the phosphorylation site often contain an essential targeting “motif” for recruitment of the responsible protein kinase isozyme.
- What is the stoichiometry of in vivo phosphorylation? Phosphorylation stoichiometry must be significant in order for this PTM to have a meaningful impact on the biological function of the target protein in a living cell.
- When is/are the particular residue(s) phosphorylated? Defining the timing and order of changes in the phosphorylation status of specific proteins following exposure to, and during recovery from, various stressors.
- How is/are the particular residue(s) phosphorylated? Pinpointing a protein’s in vivo phosphorylation site can lead to identification and characterization of the responsible protein kinase, and ultimately to the upstream signal-transduction pathways that control its expression and/or activity
- Why is the protein phosphorylated? This is perhaps the most compelling question of all, since even when Questions 1 to 4 are fully answered, we must ultimately understand how phosphorylation influences the activity and biological function(s) of the target protein.
4.3. Other PTMs
5. The Effect of Biotic and Abiotic Stresses on the Extracellular Proteome
5.1. Biotic Stresses
5.2. Abiotic Stresses
6. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of Arabidopsis thaliana. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C. Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 2004, 161, 641–675. [Google Scholar] [CrossRef]
- Jamet, E.; Canut, H.; Boudart, G.; Pont-Lezica, R.F. Cell wall proteins: A new insight through proteomics. Trends Plant Sci. 2006, 11, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Saravanan, R.S.; Damasceno, C.M.B.; Yamane, H.; Kim, B.-D.; Rose, J.K.C. Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 2004, 42, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Guimarães, L.; Pinheiro, C.; Chaves, I.; Barros, D.R.; Ricardo, C.P. Protein dynamics in the plant extracellular space. Proteomes 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Chivasa, S.; Ndimba, B.K.; Simon, W.J.; Robertson, D.; Yu, X.-L.; Knox, J.P.; Bolwell, P.; Slabas, A.R. Proteomic analysis of the Arabidopsis thaliana cell wall. Electrophoresis 2002, 23, 1754–1765. [Google Scholar] [CrossRef]
- Cassab, G.I.; Varner, J.E. Cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 321–353. [Google Scholar] [CrossRef]
- Bayer, E.M.; Bottrill, A.R.; Walshaw, J.; Vigouroux, M.; Naldrett, M.J.; Thomas, C.L.; Maule, A.J. Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 2006, 6, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Feiz, L.; Irshad, M.; Pont-Lezica, R.F.; Canut, H.; Jamet, E. Evaluation of cell wall preparations for proteomics: A new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2006, 2, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Carpin, S.; Crèvecoeur, M.; de Meyer, M.; Simon, P.; Greppin, H.; Penel, C. Identification of a Ca2+-pectate binding site on an apoplastic peroxidase. Plant Cell 2001, 13, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Kieliszewski, M.J.; Lamport, D.T.A. Extensin: Repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994, 5, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.K.; Saladié, M.; Catalá, C. The plot thickens: New perspectives of primary cell wall modification. Curr. Opin. Plant Biol. 2004, 7, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Hoson, T. Apoplast as the site of response to environmental signals. J. Plant Res. 1998, 111, 167–177. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, G.; Ferrari, S. Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 2002, 5, 295–299. [Google Scholar] [CrossRef]
- Jones, D.A.; Takemoto, D. Plant innate immunity—Direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 2004, 16, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Torii, K.U. Leucine-rich repeat receptor kinases in plants: Structure, function, and signal transduction pathways. Int. Rev. Cytol. 2004, 234, 1–46. [Google Scholar] [PubMed]
- Lertpiriyapong, K.; Sung, Z.R. The elongation defective1 mutant of Arabidopsis is impaired in the gene encoding a serine-rich secreted protein. Plant Mol. Biol. 2003, 53, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Rojo, E.; Sharma, V.K.; Kovaleva, V.; Raikhel, N.V.; Fletcher, J.C. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 2002, 14, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Von Groll, U.; Berger, D.; Altmann, T. The subtilisin-like Serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 2002, 14, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Heazlewood, J.L.; Tonti-Filippini, J.; Verboom, R.E.; Millar, A.H. Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 2005, 139, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Canut, H.; Albenne, C.; Jamet, E. Post-translational modifications of plant cell wall proteins and peptides: A survey from a proteomics point of view. Biochim. Biophys. Acta 2016, 1864, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Spiro, R.G. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002, 12, 43–56. [Google Scholar] [CrossRef]
- Faye, L.; Boulaflous, A.; Benchabane, M.; Gomord, V.; Michaud, D. Protein modifications in the plant secretory pathway: Current status and practical implications in molecular pharming. Vaccine 2005, 23, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-K.; Yokoyama, R.; Nishitani, K. A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 2005, 46, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Ndimba, B.K.; Chivasa, S.; Hamilton, J.M.; Simon, W.J.; Slabas, A.R. Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 2003, 3, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.K.C.; Bashir, S.; Giovannoni, J.J.; Jahn, M.M.; Saravanan, R.S. Tackling the plant proteome: Practical approaches, hurdles and experimental tools. Plant J. 2004, 39, 715–733. [Google Scholar] [CrossRef] [PubMed]
- Spadoni, S.; Zabotina, O.; Matteo, A.D.; Mikkelsen, J.D.; Cervone, F.; Lorenzo, G.D.; Mattei, B.; Bellincampi, D. Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of Arginine and Lysine. Plant Physiol. 2006, 141, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Brisson, L.F.; Tenhaken, R.; Lamb, C. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell 1994, 6, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Brady, J.D.; Sadler, I.H.; Fry, S.C. Di-isodityrosine, a novel tetrametric derivative of tyrosine in plant cell wall proteins: A new potential cross-link. Biochem. J. 1996, 315, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Rose, J.K.C.; Lee, S.-J. Straying off the highway: Trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. 2010, 153, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Alexandersson, E.; Ali, A.; Resjö, S.; Andreasson, E. Plant secretome proteomics. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010, 22, 4009–4030. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Kim, S.T.; Cho, W.K.; Rim, Y.; Kim, S.; Kim, S.-W.; Kang, K.Y.; Park, Z.Y.; Kim, J.-Y. Proteomics of weakly bound cell wall proteins in rice calli. J. Plant Physiol. 2009, 166, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Rajamani, U.; Verma, J.; Subba, P.; Chakraborty, N.; Datta, A.; Chakraborty, S.; Chakraborty, N. Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: A proteomic approach. J. Proteome Res. 2010, 9, 3443–3464. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Alvarez, S.; Marsh, E.L.; Lenoble, M.E.; Cho, I.-J.; Sivaguru, M.; Chen, S.; Nguyen, H.T.; Wu, Y.; Schachtman, D.P.; et al. Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol. 2007, 145, 1533–1548. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Goodger, J.Q.D.; Marsh, E.L.; Chen, S.; Asirvatham, V.S.; Schachtman, D.P. Characterization of the maize xylem sap proteome. J. Proteome Res. 2006, 5, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Chivasa, S.; Simon, W.J.; Yu, X.-L.; Yalpani, N.; Slabas, A.R. Pathogen elicitor-induced changes in the maize extracellular matrix proteome. Proteomics 2005, 5, 4894–4904. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Chen, S.; Alvarez, S.; Asirvatham, V.S.; Schachtman, D.P.; Wu, Y.; Sharp, R.E. Cell wall proteome in the maize primary root elongation zone. I. Extraction and identification of water-soluble and lightly ionically bound proteins. Plant Physiol. 2006, 140, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.-J.; Oyanagi, A.; Komatsu, S. Cell wall proteome of wheat roots under flooding stress using gel-based and LC MS/MS-based proteomics approaches. Biochim. Biophys. Acta 2010, 1804, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Houterman, P.M.; Speijer, D.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C.; Rep, M. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol. Plant Pathol. 2007, 8, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Howe, K.J.; Matas, A.J.; Buda, G.J.; Thannhauser, T.W.; Rose, J.K.C. Mining the surface proteome of tomato (Solanum lycopersicum) fruit for proteins associated with cuticle biogenesis. J. Exp. Bot. 2010, 61, 3759–3771. [Google Scholar] [CrossRef] [PubMed]
- Dahal, D.; Pich, A.; Braun, H.P.; Wydra, K. Analysis of cell wall proteins regulated in stem of susceptible and resistant tomato species after inoculation with Ralstonia solanacearum: A proteomic approach. Plant Mol. Biol. 2010, 73, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Floerl, S.; Druebert, C.; Majcherczyk, A.; Karlovsky, P.; Kües, U.; Polle, A. Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Buhtz, A.; Giavalisco, P. Analysis of xylem sap proteins from Brassica napus. BMC Plant Biol. 2005, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, B.S.; Lei, Z.; Dixon, R.A.; Sumner, L.W. Proteomics of Medicago sativa cell walls. Phytochemistry 2004, 65, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Kobayashi, Y.; Nishizawa, K.; Nanjo, Y.; Furukawa, K. Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 2010, 39, 1435–1449. [Google Scholar] [CrossRef] [PubMed]
- Gokulakannan, G.G.; Niehaus, K. Characterization of the Medicago truncatula cell wall proteome in cell suspension culture upon elicitation and suppression of plant defense. J. Plant Physiol. 2010, 167, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Dani, V.; Simon, W.J.; Duranti, M.; Croy, R.R.D. Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 2005, 5, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Pechanova, O.; Hsu, C.-Y.; Adams, J.P.; Pechan, T.; Vandervelde, L.; Drnevich, J.; Jawdy, S.; Adeli, A.; Suttle, J.C.; Lawrence, A.M.; et al. Apoplast proteome reveals that extracellular matrix contributes to multistress response in poplar. BMC Genom. 2010, 11, 674–695. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Esteso, M.J.; Sellés-Marchart, S.; Vera-Urbina, J.C.; Pedreño, M.A.; Bru-Martinez, R. Changes of defense proteins in the extracellular proteome of grapevine (Vitis vinifera cv. Gamay) cell cultures in response to elicitors. J. Proteom. 2009, 73, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Negri, A.S.; Prinsi, B.; Scienza, A.; Morgutti, S.; Cocucci, M.; Espen, L. Analysis of grape berry cell wall proteome: A comparative evaluation of extraction methods. J. Plant Physiol. 2008, 165, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, D.; Pandey, A.; Chattopadhyay, A.; Choudhary, M.K.; Chakraborty, S.; Datta, A.; Chakraborty, N. Extracellular matrix proteome of chickpea (Cicer arietinum L.) illustrates pathway abundance, novel protein functions and evolutionary perspect. J. Proteome Res. 2006, 5, 1711–1720. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, D.; Pandey, A.; Choudhary, M.K.; Datta, A.; Chakraborty, S.; Chakraborty, N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol. Cell. Proteom. 2007, 6, 1868–1884. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, D.; Jaiswal, D.K.; Ray, D.; Basu, D.; Datta, A.; Chakraborty, S.; Chakraborty, N. Dehydration-responsive reversible and irreversible changes in the extracellular matrix: Comparative proteomics of chickpea genotypes with contrasting tolerance. J. Proteome Res. 2011, 10, 2027–2046. [Google Scholar] [CrossRef] [PubMed]
- Ligat, L.; Lauber, E.; Albenne, C.; San Clemente, H.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 2011, 11, 1798–1813. [Google Scholar] [CrossRef] [PubMed]
- Day, A.; Fénart, S.; Neutelings, G.; Hawkins, S.; Rolando, C.; Tokarski, C. Identification of cell wall proteins in the flax (Linum usitatissimum) stem. Proteomics 2013, 13, 812–825. [Google Scholar] [CrossRef] [PubMed]
- Elagamey, E.; Narula, K.; Sinha, A.; Aggarwal, P.R.; Ghosh, S.; Chakraborty, N.; Chakraborty, S. Extracellular matrix proteome and phosphoproteome of potato reveals functionally distinct and diverse canonical and non-canonical proteoforms. Proteomes 2016, 4, 20–41. [Google Scholar] [CrossRef]
- Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; Silva, M.d.C.; Cardoso, F.M.H.; Planchon, S.; Barros, D.R.; Renaut, J.; Ricardo, C.P. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Calderan-Rodrigues, M.J.; Jamet, E.; Bonassi, M.B.C.R.; Guidetti-Gonzalez, S.; Begossi, A.C.; Setem, L.V.; Franceschini, L.M.; Fonseca, J.G.; Labate, C.A. Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 2014, 14, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Calderan-Rodrigues, M.J.; Jamet, E.; Douché, T.; Bonassi, M.B.R.; Cataldi, T.R.; Fonseca, J.G.; San Clemente, H.; Pont-Lezica, R.; Labate, C.A. Cell wall proteome of sugarcane stems: Comparison of a destructive and a non-destructive extraction method showed differences in glycoside hydrolases and peroxidases. BMC Plant Biol. 2016, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San Clemente, H.; Jamet, E. WallProtDB, a database resource for plant cell wall proteomics. Plant Methods 2015, 11, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Di Palma, S.; Hennrich, M.L.; Heck, A.J.R.; Mohammed, S. Recent advances in peptide separation by multidimensional liquid chromatography for proteome analysis. J. Proteom. 2012, 75, 3791–3813. [Google Scholar] [CrossRef] [PubMed]
- Charmont, S.; Jamet, E.; Pont-Lezica, R.; Canut, H. Proteomic analysis of secreted proteins from Arabidopsis thaliana seedlings: Improved recovery following removal of phenolic compounds. Phytochemistry 2005, 66, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Francis, J.L.; Whittal, R.M.; Stephens, J.L.; Wang, Y.; Zaiane, O.R.; Goebel, R.; Muench, D.G.; Good, A.G.; Taylor, G.J. Extracellular proteomes of Arabidopsis thaliana and Brassica napus roots: Analysis and comparison by MudPIT and LC-MS/MS. Plant Soil 2006, 286, 357–376. [Google Scholar] [CrossRef]
- Cheng, F.; Blackburn, K.; Lin, Y.; Goshe, M.B.; Williamson, J.D. Absolute protein quantification by LC/MSE for global analysis of salicylic acid-induced plant protein secretion responses. J. Proteome Res. 2009, 8, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Plaxton, W.C. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics 2008, 8, 4317–4326. [Google Scholar] [CrossRef] [PubMed]
- Kaffarnik, F.A.R.; Jones, A.M.E.; Rathjen, J.P.; Peck, S.C. Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol. Cell. Proteom. 2009, 8, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Borderies, G.; Jamet, E.; Lafitte, C.; Rossignol, M.; Jauneau, A.; Boudart, G.; Monsarrat, B.; Esquerré-Tugayé, M.-T.; Boudet, A.; Pont-Lezica, R. Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: A critical analysis. Electrophoresis 2003, 24, 3421–3432. [Google Scholar] [CrossRef] [PubMed]
- Haslam, R.P.; Downie, A.L.; Raveton, M.; Gallardo, K.; Job, D.; Pallett, K.E.; John, P.; Parry, M.A.J.; Coleman, J.O.D. The assessment of enriched apoplastic extracts using proteomic approaches. Ann. Appl. Biol. 2003, 143, 81–91. [Google Scholar] [CrossRef]
- Boudart, G.; Jamet, E.; Rossignol, M.; Lafitte, C.; Borderies, G.; Jauneau, A.; Esquerré-Tugayé, M.-T.; Pont-Lezica, R. Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: Identification by mass spectrometry and bioinformatics. Proteomics 2005, 5, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Casasoli, M.; Spadoni, S.; Lilley, K.S.; Cervone, F.; De Lorenzo, G.; Mattei, B. Identification by 2-D DIGE of apoplastic proteins regulated by oligogalacturonides in Arabidopsis thaliana. Proteomics 2008, 8, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Irshad, M.; Canut, H.; Borderies, G.; Pont-Lezica, R.; Jamet, E. A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: Confirmed actors and newcomers. BMC Plant Biol. 2008, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Giboulot, A.; Zivy, M.; Valot, B.; Jamet, E.; Albenne, C. Combining various strategies to increase the coverage of the plant cell wall glycoproteome. Phytochemistry 2011, 72, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Minic, Z.; Jamet, E.; Négroni, L.; der Garabedian, P.A.; Zivy, M.; Jouanin, L. A sub-proteome of Arabidopsis thaliana mature stems trapped on Concanavalin A is enriched in cell wall glycoside hydrolases. J. Exp. Bot. 2007, 58, 2503–2512. [Google Scholar] [CrossRef] [PubMed]
- Borner, G.H.H.; Lilley, K.S.; Stevens, T.J.; Dupree, P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 2003, 132, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Schultz, C.J.; Ferguson, K.L.; Lahnstein, J.; Bacic, A. Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J. Biol. Chem. 2004, 279, 45503–45511. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.; Mitchell, G.P.; Gilroy, J.S.; Gerrish, C.; Bolwell, G.P.; Slabas, A.R. Differential extraction and protein sequencing reveals major differences in patterns of primary cell wall proteins from plants. J. Biol. Chem. 1997, 272, 15841–15848. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Song, Y. Differential proteomic analysis of apoplastic proteins during initial phase of salt stress in rice. Plant Signal. Behav. 2009, 4, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, D.D.; Duff, S.M.G.; Fife, C.A.; Julien-Inalsingh, C.; Plaxton, W.C. Response to phosphate deprivation in Brassica nigra suspension cells. Plant Physiol. 1990, 93, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Bozzo, G.G.; Dunn, E.L.; Plaxton, W.C. Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ. 2006, 29, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Deracinois, B.; Flahaut, C.; Duban-Deweer, S.; Karamanos, Y. Comparative and quantitative global proteomics approaches: An overview. Proteomes 2013, 1, 180–218. [Google Scholar] [CrossRef]
- Hennrich, M.L.; Gavin, A.-C. Quantitative mass spectrometry of posttranslational modifications: Keys to confidence. Sci. Signal. 2015, 8, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Nikolovski, N.; Shliaha, P.V.; Gatto, L.; Dupree, P.; Lilley, K.S. Label-free protein quantification for plant Golgi protein localization and abundance. Plant Physiol. 2014, 166, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Helm, S.; Dobritzsch, D.; Rödiger, A.; Agne, B.; Baginsky, S. Protein identification and quantification by data-independent acquisition and multi-parallel collision-induced dissociation mass spectrometry (MSE) in the chloroplast stroma proteome. J. Proteomics 2014, 98, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Qian, W.; Hurley, B.A.; She, Y.-M.; Wang, D.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 2010, 33, 1789–1803. [Google Scholar] [CrossRef] [PubMed]
- Robinson, W.D.; Park, J.; Tran, H.T.; Del Vecchio, H.A.; Ying, S.; Patel, K.; McKnight, T.D.; Plaxton, W.C. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate scavenging by Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 6531–6542. [Google Scholar] [CrossRef] [PubMed]
- Veljanovski, V.; Vanderbeld, B.; Knowles, V.L.; Snedden, W.A.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 2006, 142, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Tsiatsiani, L.; Heck, A.J.R. Proteomics beyond trypsin. FEBS J. 2015, 282, 2612–2626. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.-W.; Mohanty, A.; Chary, S.N.; Li, S.; Paap, B.; Drakakaki, G.; Kopec, C.D.; Li, J.; Ehrhardt, D.; Jackson, D.; et al. High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 2004, 135, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Koroleva, O.A.; Tomlinson, M.L.; Leader, D.; Shaw, P.; Doonan, J.H. High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 2005, 41, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Rose, J.K.C. A yeast secretion trap assay for identification of secreted proteins from eukaryotic phytopathogens and their plant hosts. Methods Mol. Biol. 2012, 835, 519–530. [Google Scholar] [PubMed]
- Groover, A.T.; Fontana, J.R.; Arroyo, J.M.; Yordan, C.; McCombie, W.R.; Martienssen, R.A. Secretion trap tagging of secreted and membrane-spanning proteins using Arabidopsis gene traps. Plant Physiol. 2003, 132, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.A.; Collins-Racie, L.A.; Colbert, M.; Duckett, M.; Golden-Fleet, M.; Kelleher, K.; Kriz, R.; LaVallie, E.R.; Merberg, D.; Spaulding, V.; et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene 1997, 198, 289–296. [Google Scholar] [CrossRef]
- Kristoffersen, P.; Teichmann, T.; Stracke, R.; Palme, K. Signal sequence trap to clone cDNAs encoding secreted or membrane-associated plant proteins. Anal. Biochem. 1996, 243, 127–132. [Google Scholar] [CrossRef]
- Gomord, V.; Faye, L. Posttranslational modification of therapeutic proteins in plants. Curr. Opin. Plant Biol. 2004, 7, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Jensen, O.N. Modification-specific proteomics: Characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004, 8, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Elbers, I.J.; Stoopen, G.M.; Bakker, H.; Stevens, L.H.; Bardor, M.; Molthoff, J.W.; Jordi, W.J.; Bosch, D.; Lommen, A. Influence of growth conditions and developmental stage on N-glycan heterogeneity of transgenic immunoglobulin G and endogenous proteins in tobacco leaves. Plant Physiol. 2001, 126, 1314–1322. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, P.; Cabanes-Macheteau, M.; Rayon, C.; Fischette-Lainé, A.-C.; Gomord, V.; Faye, L. N-Glycoprotein biosynthesis in plants: Recent developments and future trends. Plant Mol. Biol. 1998, 38, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Van Ree, R.; Cabanes-Macheteau, M.; Akkerdaas, J.; Milazzo, J.P.; Loutelier-Bourhis, C.; Rayon, C.; Villalba, M.; Koppelman, S.; Aalberse, R.; Rodriguez, R.; et al. Beta(1,2)-xylose and alpha(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J. Biol. Chem. 2000, 275, 11451–11458. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, H.A.; Ying, S.; Park, J.; Knowles, V.L.; Kanno, S.; Tanoi, K.; She, Y.-M.; Plaxton, W.C. The cell wall-targeted purple acid phosphatase AtPAP25 is critical for acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation. Plant J. 2014, 80, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Shane, M.W. The role of post-translational enzyme modifications in the metabolic adaptations of phosphorus-deprived plants. In Annual Plant Reviews; Plaxton, W.C., Lambers, H., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 2015; Volume 48, pp. 99–123. [Google Scholar]
- Chrispeels, M.J.; Raikhel, N.V. Lectins, lectin genes, and their role in plant defense. Plant Cell 1991, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- De Moreira, R.A.; Ainouz, I.L.; De Oliveira, J.T.; Cavada, B.S. Plant lectins, chemical and biological aspects. Mem. Inst. Oswaldo Cruz 1991, 86, 211–218. [Google Scholar] [CrossRef]
- Velasquez, S.M.; Ricardi, M.M.; Dorosz, J.G.; Fernandez, P.V.; Nadra, A.D.; Pol-Fachin, L.; Egelund, J.; Gille, S.; Harholt, J.; Ciancia, M.; et al. O-glycosylated cell wall proteins are essential in root hair growth. Science 2011, 332, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Hall, Q.; Cannon, M.C. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 2002, 14, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Gille, S.; Hänsel, U.; Ziemann, M.; Pauly, M.; Chrispeels, M.J. Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. USA 2009, 106, 14699–14704. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.C. Exploring the role of protein phosphorylation in plants: From signalling to metabolism. Biochem. Soc. Trans. 2007, 35, 28–32. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente van Bentem, S.; Hirt, H. Protein tyrosine phosphorylation in plants: More abundant than expected? Trends Plant Sci. 2009, 14, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Moorhead, G.B.G.; Templeton, G.W.; Tran, H.T. Role of protein kinases, phosphatases and 14-3-3 proteins in the control of primary plant metabolism. In Annual Plant Reviews Volume 22: Control of Primary Metabolism in Plants; Plaxton, W.C., McManus, M.T., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 121–149. [Google Scholar]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Tagliabracci, V.S.; Pinna, L.A.; Dixon, J.E. Secreted protein kinases. Trends Biochem. Sci. 2013, 38, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef] [PubMed]
- Mazzucotelli, E.; Mastrangelo, A.M.; Crosatti, C.; Guerra, D.; Stanca, A.M.; Cattivelli, L. Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Sci. 2008, 174, 420–431. [Google Scholar] [CrossRef]
- Vierstra, R.D. The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol. 2012, 160, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, M.; Suryadinata, R.; Tan, A.R.; Roesley, S.N.A.; Sarcevic, B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life 2012, 64, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, R.G.; She, Y.-M.; Leach, C.A.; Plaxton, W.C. Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J. Biol. Chem. 2008, 283, 29650–29657. [Google Scholar] [CrossRef] [PubMed]
- Shane, M.W.; Fedosejevs, E.T.; Plaxton, W.C. Reciprocal control of anaplerotic phosphoenolpyruvate carboxylase by in vivo monoubiquitination and phosphorylation in developing proteoid roots of phosphate-deficient harsh hakea. Plant Physiol. 2013, 161, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Schnell, J.D.; Hicke, L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003, 278, 35857–35860. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, D.; Riezman, H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007, 315, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Hu, Z.; Lü, H.; Wang, X.; Baluska, F.; Samaj, J.; Lin, J. Roles of the ubiquitin/proteasome pathway in pollen tube growth with emphasis on MG132-induced alterations in ultrastructure, cytoskeleton, and cell wall components. Plant Physiol. 2006, 141, 1578–1590. [Google Scholar] [CrossRef] [PubMed]
- Rogers, L.D.; Overall, C.M. Proteolytic post-translational modification of proteins: Proteomic tools and methodology. Mol. Cell. Proteom. 2013, 12, 3532–3542. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Rausch, T.; Greiner, S. The N-terminal pro region mediates retention of unprocessed type-I PME in the Golgi apparatus. Plant J. 2009, 58, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Hurley, B.A.; Tran, H.T.; Marty, N.J.; Park, J.; Snedden, W.A.; Mullen, R.T.; Plaxton, W.C. The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol. 2010, 153, 1112–1122. [Google Scholar] [CrossRef] [PubMed]
- Hamann, T. Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, T.V.; Bonetta, D.T.; Goring, D.R. Sentinels at the wall: Cell wall receptors and sensors. New Phytol. 2007, 176, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Hématy, K.; Cherk, C.; Somerville, S. Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 2009, 12, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Ringli, C. Monitoring the outside: Cell wall-sensing mechanisms. Plant Physiol. 2010, 153, 1445–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, G.J.; Blaukopf, C. Irritable walls: The plant extracellular matrix and signaling. Plant Physiol. 2010, 153, 467–478. [Google Scholar] [CrossRef]
- Stress signaling in plants: Genomics and proteomics perspective. In Stress Signaling in Plants: Genomics and Proteomics Perspective; Sarwat, M.; Ahmad, A.; Abdin, M. (Eds.) Springer: New York, NY, USA, 2013; Volume 1.
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, G.; D’Ovidio, R.; Cervone, F. The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 2001, 39, 313–335. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, S.-Y.; Zhao, W.-S.; Su, S.-C.; Peng, Y.-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant. Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Marchand, C.; Le Maréchal, P.; Meyer, Y.; Miginiac-Maslow, M.; Issakidis-Bourguet, E.; Decottignies, P. New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 2004, 4, 2696–2706. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bindschedler, L.V.; Blee, K.A.; Butt, V.S.; Davies, D.R.; Gardner, S.L.; Gerrish, C.; Minibayeva, F. The apoplastic oxidative burst in response to biotic stress in plants: A three-component system. J. Exp. Bot. 2002, 53, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Pogorelko, G.; Lionetti, V.; Fursova, O.; Sundaram, R.M.; Qi, M.; Whitham, S.A.; Bogdanove, A.J.; Bellincampi, D.; Zabotina, O.A. Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 2013, 162, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Decreux, A.; Messiaen, J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005, 46, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Zagorchev, L.; Kamenova, P.; Odjakova, M.; Zagorchev, L.; Kamenova, P.; Odjakova, M. The role of plant cell wall proteins in response to salt stress. Sci. World J. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, J.M.; Brumer, H. The XTH gene family: An update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol. 2010, 153, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Rienth, M.; Torregrosa, L.; Luchaire, N.; Chatbanyong, R.; Lecourieux, D.; Kelly, M.T.; Romieu, C. Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigeto, J.; Kiyonaga, Y.; Fujita, K.; Kondo, R.; Tsutsumi, Y. Putative cationic cell-wall-bound peroxidase homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J. Agric. Food Chem. 2013, 61, 3781–3788. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-R.; Kim, K.-Y.; Jung, W.-J.; Avice, J.-C.; Ourry, A.; Kim, T.-H. Peroxidases and lignification in relation to the intensity of water-deficit stress in white clover (Trifolium repens L.). J. Exp. Bot. 2007, 58, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-A.; Shin, J.-W.; Liu, Z.-H. Effect of light on peroxidase and lignin synthesis in mungbean hypocotyls. Plant Physiol. Biochem. 2002, 40, 33–39. [Google Scholar] [CrossRef]
- Csiszár, J.; Gallé, A.; Horváth, E.; Dancsó, P.; Gombos, M.; Váry, Z.; Erdei, L.; Györgyey, J.; Tari, I. Different peroxidase activities and expression of abiotic stress-related peroxidases in apical root segments of wheat genotypes with different drought stress tolerance under osmotic stress. Plant Physiol. Biochem. 2012, 52, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Šimonovičová, M.; Huttová, J.; Mistrík, I.; Široká, B.; Tamás, L. Peroxidase mediated hydrogen peroxide production in barley roots grown under stress conditions. Plant Growth Regul. 2004, 44, 267–275. [Google Scholar] [CrossRef]
- Csiszár, J.; Pintér, B.; Kolbert, Z.; Erdei, L.; Tari, I. Peroxidase activities in root segments of wheat genotypes under osmotic stress. Acta Biol. Szeged. 2008, 52, 155–156. [Google Scholar]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5. [Google Scholar] [CrossRef] [PubMed]

| Type of Proteome | Proteome Source | Method Employed | Reference | |
|---|---|---|---|---|
| Non-Destructive: | Secretome | Culture medium of liquid-cultured seedlings | Collection of culture filtrate | [64] |
| Secretome | Culture medium of suspension cell cultures | Collection of culture filtrate | [65] | |
| Secretome | Culture medium of hydroponically-grown plants | Collection of culture filtrate | [66] | |
| Secretome | Culture medium of suspension cells treated with salicylic acid | Collection of culture filtrate | [67] | |
| Subset of the secretome | Culture medium of suspension cells grown with or without phosphate | Collection of culture filtrate | [68] | |
| Subset of the secretome | Culture medium of suspension cells infected with Pseudomonas syringae | Collection of culture filtrate | [69] | |
| Cell wall proteome | Suspension cell culture | Sequential washing of intact cells with salt solutions | [68,70] | |
| Cell wall proteome | Suspension cell culture | Wash intact cells with salt solution | [69] | |
| Apoplastic fluid proteome | Leaves | Vacuum infiltration | [71,72] | |
| Subset of the apoplastic fluid proteome | Seedlings treated with oligogalacturonides | Vacuum infiltration | [73] | |
| Destructive: | Cell wall proteome | Suspension cell culture | Salt extraction of purified cell walls | [8] |
| Cell wall proteome | Suspension cell culture | Salt extraction of purified cell walls | [6] | |
| Cell wall proteome | Etiolated hypocotyls | Salt extraction of purified cell walls | [74] | |
| Cell wall proteome | Etiolated hypocotyls | Salt extraction of purified cell walls | [9] | |
| Subset of cell wall proteome | Suspension cells treated with fungal elicitors | Salt extraction of purified cell walls | [26] | |
| Cell wall glycoproteome | Etiolated hypocotyls | Lectin affinity chromatography | [75] | |
| N-glycoproteome | Mature stems | Lectin affinity chromatography | [76] | |
| Glycosylphosphatidyl- inositol anchored proteome | Callus cells | Phospholipase C treatment of purified membrane fraction | [77] | |
| Arabinogalactan proteins proteome | Liquid-cultured etiolated seedlings | Yariv precipitation | [78] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghahremani, M.; Stigter, K.A.; Plaxton, W. Extraction and Characterization of Extracellular Proteins and Their Post-Translational Modifications from Arabidopsis thaliana Suspension Cell Cultures and Seedlings: A Critical Review. Proteomes 2016, 4, 25. https://doi.org/10.3390/proteomes4030025
Ghahremani M, Stigter KA, Plaxton W. Extraction and Characterization of Extracellular Proteins and Their Post-Translational Modifications from Arabidopsis thaliana Suspension Cell Cultures and Seedlings: A Critical Review. Proteomes. 2016; 4(3):25. https://doi.org/10.3390/proteomes4030025
Chicago/Turabian StyleGhahremani, Mina, Kyla A. Stigter, and William Plaxton. 2016. "Extraction and Characterization of Extracellular Proteins and Their Post-Translational Modifications from Arabidopsis thaliana Suspension Cell Cultures and Seedlings: A Critical Review" Proteomes 4, no. 3: 25. https://doi.org/10.3390/proteomes4030025





