Subcellular Proteomics: Application to Elucidation of Flooding-Response Mechanisms in Soybean
Abstract
:1. Introduction
2. Strengths and Problems of Subcellular Proteomic Techniques
2.1. Gel-Free and Gel-Based Techniques
2.2. Label-Free and Label-Based Techniques in Gel-Free Proteomics
3. Purification Techniques for Subcellular Proteins from Plants
3.1. Nuclei
3.2. Mitochondria
3.3. Endoplasmic Reticulum
3.4. Cell Wall
3.5. Plasma Membrane
3.6. Validation of Purity
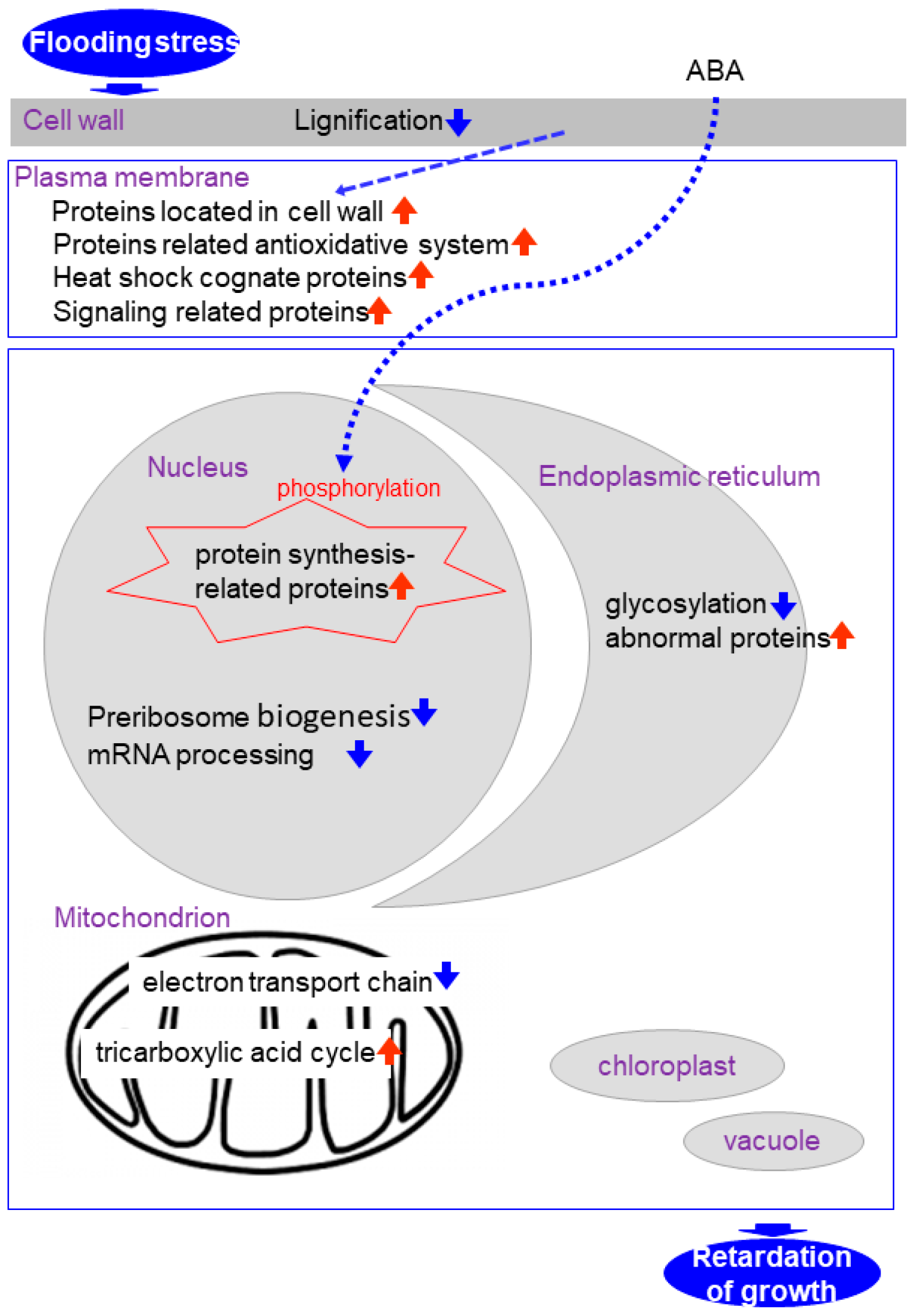
4. Subcellular Proteomics in Soybean under Flooding Stress
4.1. Nuclear Proteomics in Soybean under Flooding Stress
4.2. Mitochondrial Proteomics in Soybean under Flooding Stress
4.3. Endoplasmic Reticulum Proteomics in Soybean under Flooding Stress
4.4. Cell Wall and Plasma Membrane Proteomics in Soybean under Flooding Stress
5. Construction of a Subcellular Proteomics Related Database
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LC | liquid chromatography |
| MS | mass spectrometry |
| UPR | unfolded protein response |
References
- Hou, F.F.; Thseng, F.S. Studies on the flooding tolerance of soybean seed: Varietal differences. Euphytica 1991, 57, 169–173. [Google Scholar] [CrossRef]
- Githiri, S.M.; Watanabe, S.; Harada, K.; Takahashi, R. QTL analysis of flooding tolerance in soybean at an early vegetative growth stage. Plant Breed. 2006, 125, 613–618. [Google Scholar] [CrossRef]
- Nakayama, N.; Hashmoto, S.; Shimada, S.; Takahashi, M.; Kim, Y.H.; Oya, T.; Arihara, J. The effect of flooding stress at the germination stage on the growth of soybean in relation to initial seed moisture content. J. Crop. Sci. 2004, 73, 323–329. [Google Scholar] [CrossRef]
- Voesenek, L.A.; Colmer, T.D.; Pierik, R.; Millenaar, F.F.; Peeters, A.J. How plants cope with complete submergence. New Phytol. 2006, 170, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.A.; Wong, D.M.L.; Sachs, M.M. The anaerobic response of soybean. Plant Physiol. 1990, 92, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Greenway, H.; Colmer, T.D.; Millar, A.H. Protein synthesis by rice coleoptiles during prolonged anoxia: Implications for glycolysis, growth and energy utilization. Ann. Bot. 2005, 96, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Mutava, R.N.; Prince, S.J.; Syed, N.H.; Song, L.; Valliyodan, B.; Chen, W.; Nguyen, H.T. Understanding abiotic stress tolerance mechanisms in soybean: A comparative evaluation of soybean response to drought and flooding stress. Plant Physiol. Biochem. 2015, 86, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteom. Res. 2012, 11, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, A.; Komatsu, S. Impact of post-translational modifications of crop proteins under abiotic stress. Proteomes 2016, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Komatsu, S. Quantitative proteomics reveals the effect of protein glycosylation in soybean root under flooding stress. Front. Plant Sci. 2014, 5, 627. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Komatsu, S. Gel-free/label-free proteomic analysis of endoplasmic reticulum proteins in soybean root tips under flooding and drought stresses. J. Proteom. Res. 2016, 15, 2211–2227. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Komatsu, S. Ubiquitin/proteasome-mediated proteolysis is involved in the response to flooding stress in soybean roots, independent of oxygen limitation. Plant Sci. 2012, 185–186, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, Y.; Skultety, L.; Ashraf, Y.; Komatsu, S. Comparative proteomic analysis of early-stage soybean seedlings responses to flooding by using gel and gel-free techniques. J. Proteom. Res. 2010, 9, 3989–4002. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, Y.; Skultety, L.; Uváčková, L.; Klubicová, K.; Hajduch, M.; Komatsu, S. Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteom. Res. 2012, 11, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Sakata, K.; Komatsu, S. Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J. Proteom. Res. 2014, 13, 5618–5634. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Komatsu, S. Quantitative proteomics of nuclear phosphoproteins in the root tip of soybean during the initial stages of flooding stress. J. Proteom. 2015, 119, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Komatsu, S. Plant subcellular proteomics: Application for exploring optimal cell function in soybean. J. Proteom. 2016, 143, 45–46. [Google Scholar] [CrossRef] [PubMed]
- Hossain, Z.; Nouri, M.Z.; Komatsu, S. Plant Cell organelle proteomics in response to abiotic stress. J. Proteom. Res. 2012, 11, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Komatsu, S. Nuclear proteomics reveals the role of protein synthesis and chromatin structure in root tip of soybean during the initial stage of flooding stress. J. Proteom. Res. 2016, 15, 2283–2298. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Hiraga, S.; Nouri, M.Z. Analysis of flooding-responsive proteins localized in the nucleus of soybean root tips. Mol. Biol. Rep. 2014, 41, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yanagawa, Y. Cell wall proteomics of crop. Front. Plant Sci. 2013, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.W.; Nanjo, Y.; Komatsu, S. Identification of nuclear proteins in soybean under flooding stress using proteomic technique. Protein Pept. Lett. 2014, 21, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.H.; Komatsu, S. Involvement of reactive oxygen species and mitochondrial proteins in biophoton emission in roots of soybean plants under flooding stress. J. Proteom. Res. 2015, 14, 2219–2236. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteom. Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Kuji, R.; Nanjo, Y.; Hiraga, S.; Furukawa, K. Comprehensive analysis of endoplasmic reticulum-enriched fraction in root tips of soybean under flooding stress using proteomics techniques. J. Proteom. 2012, 77, 531–560. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Kobayashi, Y.; Nishizawa, K.; Nanjo, Y.; Furukawa, K. Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 2010, 39, 1435–1449. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Wada, T.; Abaléa, Y.; Nouri, M.Z.; Nanjo, Y.; Nakayama, N.; Shimamura, S.; Yamamoto, R.; Nakamura, T.; Furukawa, K. Analysis of plasma membrane proteome in soybean and application to flooding stress response. J. Proteom. Res. 2009, 8, 4487–4499. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [PubMed]
- Arruda, S.C.; Barbosa Hde, S.; Azevedo, R.A.; Arruda, M.A. Two-dimensional difference gel electrophoresis applied for analytical proteomics: Fundamentals and applications to the study of plant proteomics. Analyst 2011, 136, 4119–4126. [Google Scholar] [CrossRef] [PubMed]
- Vadivel, A.K.A. Gel-based proteomics in plants: Time to move on from the tradition. Front. Plant Sci. 2015, 6, 369. [Google Scholar]
- León, I.R.; Schwämmle, V.; Jensen, O.N.; Sprenger, R.R. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol. Cell. Proteom. 2013, 12, 2992–3005. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T. Membrane proteins and proteomics: Love is possible, but so difficult. Electrophoresis 2009, 30, S174–S180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barua, P.; Subba, P.; Lande, N.V.; Mangalaparthi, K.K.; Prasad, T.S.K.; Chakraborty, S.; Chakraborty, N. Gel-based and gel-free search for plasma membrane proteins in chickpea (Cicer arietinum L.) augments the comprehensive data sets of membrane protein repertoire. J. Proteom. 2016, 143, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, R. Shotgun proteomics of the barley seed proteome. BMC Genom. 2017, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Min, C.W.; Lee, S.H.; Cheon, Y.E.; Han, W.Y.; Ko, J.M.; Kang, H.W.; Kim, Y.C.; Agrawal, G.K.; Rakwal, R.; Gupta, R.; Kim, S.T. In-depth proteomic analysis of Glycine max seeds during controlled deterioration treatment reveals a shift in seed metabolism. J. Proteom. 2017, 169, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez de Francisco, L.; Romero-Rodríguez, M.C.; Navarro-Cerrillo, R.M.; Miniño, V.; Perdomo, O.; Jorrín-Novo, J.V. Characterization of the orthodox Pinus occidentalis seed and pollen Proteomes by using complementary gel-based and gel-free approaches. J. Proteom. 2016, 143, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Takáč, T.; Pechan, T.; Samajová, O.; Samaj, J. Vesicular trafficking and stress response coupled to PI3K inhibition by LY294002 as revealed by proteomic and cell biological analysis. J. Proteom. Res. 2013, 12, 4435–4448. [Google Scholar] [CrossRef] [PubMed]
- Dayon, L.; Sanchez, J.C. Relative protein quantification by MS/MS using the tandem mass tag technology. Methods Mol. Biol. 2012, 893, 115–127. [Google Scholar] [PubMed]
- Rauniyar, N.; Yates, J.R., III. Isobaric Labeling-Based Relative Quantification in Shotgun Proteomics. J. Proteom. Res. 2014, 13, 5293–5309. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Cai, J.; Zhang, Y.; Qin, G.; Tian, S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol. 2014, 15, 548. [Google Scholar] [CrossRef] [PubMed]
- Ford, K.L.; Chin, T.; Srivastava, V.; Zeng, W.; Doblin, M.S.; Bulone, V.; Bacic, A. Comparative “Golgi” proteome study of Lolium multiflorum and Populus trichocarpa. Proteomes 2016, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ding, X.; Han, S.; He, T.; Zhang, H.; Yang, L.; Yang, S.; Gai, J. Differential proteomics analysis to identify proteins and pathways associated with male sterility of soybean using iTRAQ-based strategy. J Proteom. 2016, 138, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Neilson, K.A.; Ali, N.A.; Muralidharan, S.; Mirzaei, M.; Mariani, M.; Assadourian, G.; Lee, A.; van Sluyter, S.C.; Haynes, P.A. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics 2011, 11, 535–553. [Google Scholar] [CrossRef] [PubMed]
- Tsou, C.C.; Tsai, C.F.; Tsui, Y.H.; Sudhir, P.R.; Wang, Y.T.; Chen, Y.J.; Chen, J.Y.; Sung, T.Y.; Hsu, W.L. IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol. Cell. Proteom. 2010, 9, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Sakata, K.; Ohyanagi, H.; Nobori, H.; Nakamura, T.; Hashiguchi, A.; Nanjo, Y.; Mikami, Y.; Yunokawa, H.; Komatsu, S. Soybean proteome database: A data resource for plant differential omics. J. Proteom. Res. 2009, 8, 3539–3548. [Google Scholar] [CrossRef] [PubMed]
- Meier, I.; Richards, E.J.; Evans, D.E. Cell biology of the plant nucleus. Annu. Rev. Plant Biol. 2017, 68, 139–172. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S. Isolation, purity assessment, and proteomic analysis of nuclei. Methods Mol. Biol. 2018, 1696, 1681–1690. [Google Scholar]
- Smith-Hammond, C.L.; Hoyos, E.; Miernyk, J.A. The pea seedling mitochondrial Nε-lysine acetylome. Mitochondrion 2014, 19, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Parsons, H.T. Preparation of Highly Enriched ER Membranes Using Free-Flow Electrophoresis. Methods Mol. Biol. 2018, 1691, 103–115. [Google Scholar] [PubMed]
- Phillips, M.J.; Voeltz, G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016, 17, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Eubel, H.; O’Toole, N.; Millar, A.H. Combining proteomics of root and shoot mitochondria and transcript analysis to define constitutive and variable components in plant mitochondria. Phytochemistry 2011, 72, 1092–1108. [Google Scholar] [CrossRef] [PubMed]
- Keegstra, K. Plant Cell walls. Plant Physiol. 2010, 154, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Petrovská, B.; Jeřábková, H.; Chamrád, I.; Vrána, J.; Lenobel, R.; Uřinovská, J.; Sebela, M.; Doležel, J. Proteomic analysis of barley cell nuclei purified by flow sorting. Cytogenet. Genome Res. 2014, 143, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Taylor, N.L.; Narsai, R.; Eubel, H.; Whelan, J.; Millar, A.H. Experimental analysis of the rice mitochondrial proteome, its biogenesis, and heterogeneity. Plant Physiol. 2009, 149, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Kriechbaumer, V.; Botchway, S.W.; Slade, S.E.; Knox, K.; Frigerio, L.; Oparka, K.; Hawes, C. Reticulomics: Protein-protein interaction studies with two plasmodesmata-localized reticulon family proteins identify binding partners enriched at plasmodesmata, endoplasmic reticulum, and the plasma membrane. Plant Physiol. 2015, 169, 1933–1945. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.K.; Walters, B.T.; Clouse, S.D.; Goshe, M.B. An efficient organic solvent based extraction method for the proteomic analysis of Arabidopsis plasma membranes. J. Proteom. Res. 2009, 8, 2752–2767. [Google Scholar] [CrossRef] [PubMed]
- De Michele, R.; McFarlane, H.E.; Parsons, H.T.; Meents, M.J.; Lao, J.; González Fernández-Niño, S.M.; Petzold, C.J.; Frommer, W.B.; Samuels, A.L.; Heazlewood, J.L. Free-flow electrophoresis of plasma membrane vesicles enriched by two-phase partitioning enhances the quality of the proteome from Arabidopsis seedlings. J. Proteom. Res. 2016, 15, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Hamada, S.; Hiratsuka, M.; Fukao, Y.; Kawasaki, T.; Shimamoto, K. Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol. 2009, 50, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Tanner, K.G.; Denu, J.M. A continuous, nonradioactive assay for histone acetyltransferases. Anal. Biochem. 2000, 280, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Lisenbee, C.S.; Lingard, M.J.; Trelease, R.N. Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 2005, 43, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Haslbeck, M.; Babujee, L.; Jahn, O.; Reumann, S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006, 141, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.J.; Cosgrove, D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000, 51, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Saravanan, R.S.; Damascenom, C.M.B.; Yamane, H.; Kim, B.D.; Rose, J.K.C. Digging deeper into the Plant Cell wall proteome. Plant Physiol. Biochem. 2004, 42, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Calderan-Rodrigues, M.J.; Jamet, E.; Bonassi, M.B.; Guidetti-Gonzalez, S.; Begossi, A.C.; Setem, L.V.; Franceschini, L.M.; Fonseca, J.G.; Labate, C.A. Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 2014, 14, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Duruflé, H.; Clemente, H.S.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 2017, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hervé, V.; Duruflé, H.; San Clemente, H.; Albenne, C.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. An enlarged cell wall proteome of Arabidopsis thaliana rosettes. Proteomics 2016, 16, 3183–3187. [Google Scholar] [CrossRef] [PubMed]
- Santoni, V. Plant plasma membrane protein extraction and solubilization for proteomic analysis. Methods Mol. Biol. 2007, 355, 93–109. [Google Scholar] [PubMed]
- Malinsky, J.; Opekarová, M.; Grossmann, G.; Tanner, W. Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu Rev Plant Biol. 2013, 64, 501–529. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; He, L.F.; Sasaki, T.; Yamamoto, Y.; Zheng, S.J.; Ligaba, A.; Yan, X.L.; Ahn, S.J.; Yamaguchi, M.; Sasakawa, H.; et al. Citrate secretion coupled with the modulation of soybean root tip under aluminum stress. Up-regulation of transcription, translation, and threonine-oriented phosphorylation of plasma membrane H+-ATPase. Plant Physiol. 2005, 138, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Reumann, S.; Babujee, L.; Ma, C.; Wienkoop, S.; Siemsen, T.; Antonicelli, G.E.; Rasche, N.; Lüder, F.; Weckwerth, W.; Jahn, O. Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 2007, 19, 3170–3193. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Plant mitochondrial proteomics. In Plant Proteomics: Methods and Protocol; Jorrin-Novo, J.V., Komatsu, S., Weckwerth, W., Wienkoop, S., Eds.; Springer: New York, NY, USA, 2014; pp. 499–525. [Google Scholar]
- Gomez, L.; Chrispeels, M.J. Complementation of an Arabidopsis thaliana mutant that lacks complex asparagine-linked glycans with the human cDNA encoding N-acetylglucosaminyltransferase I. Proc. Natl. Acad. Sci. USA 1994, 91, 1829–1833. [Google Scholar] [CrossRef] [PubMed]
- Tugal, H.B.; Pool, M.; Baker, A. Arabidopsis 22-kilodalton peroxisomal membrane protein. Nucleotide sequence analysis and biochemical characterization. Plant Physiol. 1999, 120, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.J.; Oyanagi, A.; Komatsu, S. Cell wall proteome of wheat roots under flooding stress using gel-based and LC MS/MS-based proteomics approaches. Biochim. Biophys. Acta 2010, 1804, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Palm, D.; Simm, S.; Darm, K.; Weis, B.L.; Ruprecht, M.; Schleiff, E.; Scharf, C. Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol. 2016, 13, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumar, A.; Subba, P.; Gayali, S.; Barua, P.; Chakraborty, S.; Chakraborty, N. Nuclear phosphoproteome of developing chickpea seedlings (Cicer arietinum L.) and protein-kinase interaction network. J. Proteom. 2014, 105, 58–73. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Alegre, S.; Nagler, M.; Escandón, M.; Annacondia, M.L.; Weckwerth, W.; Valledor, L.; Cañal, M.J. The variations in the nuclear proteome reveal new transcription factors and mechanisms involved in UV stress response in Pinus radiata. J. Proteom. 2016, 143, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Fujiwara, M.; Furuto, A.; Fukao, Y.; Yamashita, T.; Kamo, M.; Kawamura, Y.; Uemura, M. Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 2009, 50, 341–359. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, D.; Kawamura, Y.; Uemura, M. Changes of detergent-resistant plasma membrane proteins in oat and rye during cold acclimation: Association with differential freezing tolerance. J. Proteom. Res. 2013, 12, 4998–5011. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteom. Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.; Greenway, H. Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol. 2003, 30, 1–47. [Google Scholar] [CrossRef]
- Mustafa, G.; Komatsu, S. Insights into the response of soybean mitochondrial proteins to various sizes of aluminum oxide nanoparticles under flooding stress. J. Proteom. Res. 2016, 15, 4464–4475. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Wall structure and wall loosening. A look backwards and forwards. Plant Physiol. 2001, 125, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; Ribeiro, J.M.; Vatulescu, A.D.; Findlay, K.; MacDougall, A.J.; Jackson, P.A. Extensin network formation in Vitis vinifera callus cells is an essential and causal event in rapid and H2O2 induced reduction in primary cell wall hydration. BMC Plant Biol. 2011, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Tournaire-Roux, C.; Sutka, M.; Javot, H.; Gout, E.; Gerbeau, P.; Luu, D.-T.; Bllgny, R.; Maurel, C. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaoirins. Nature 2003, 425, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Ohyanagi, H.; Sakata, K.; Komatsu, S. Soybean Proteome Database 2012: Update on the comprehensive data repository for soybean proteomics. Front. Plant Sci. 2012, 3, 110. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Wang, X.; Yin, X.; Nanjo, Y.; Ohyanagi, H.; Sakata, K. Integration of gel-based and gel-free proteomic data for functional analysis of proteins through Soybean Proteome Database. J. Proteom. 2017, 163, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, M.; Kanamori, H.; Komatsu, S.; Namiki, N.; Mukai, Y.; Kurita, K.; Kamatsuki, K.; Ikawa, H.; Yano, R.; Ishimoto, M.; et al. The Glycine max cv. Enrei genome for improvement of Japanese soybean cultivars. Int. J. Genom. 2015, 2015, 358127. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Pouya, F.M.; Ghaffari, M.R.; Mirzaei, M.; Ghaffari, A.; Alikhani, M.; Ghareyazie, M.; Komatsu, S.; Haynes, P.A.; Salekdeh, G.H. PlantPReS: A database for plant proteome response to stress. J. Proteom. 2016, 143, 69–72. [Google Scholar] [CrossRef] [PubMed]
| Organ | Species | Purification | Proteome Analysis | Identified Proteins | Representative Ref |
|---|---|---|---|---|---|
| Nucleus | |||||
| Cell culture | Arabidopsis | Density gradient | LC-MS/MS | 2544 proteins | [54] |
| Aerial parts | Chickpea | Density gradient | 2DE LC-ESI-MS/MS, MALDI-TOF/TOF | 107 phosphoproteins | [52] |
| Seedlings | Pinus radiata | Density gradient | LTQ-Orbitrap MS | 33 transcription factors/regulators | [55] |
| Grains | Barley | Flow cytometric sorting | 1DE LC-MS/MS MALDI-MS/MS | 803 nuclear proteins | [49] |
| Mitochondrion | |||||
| Shoot | Rice | Density gradient, Free-flow electrophoresis | Gel based/LC-MS/MS | 322 proteins | [56] |
| Seedlings | Pea | Density gradient | LC–MS/MS | 358 Lys-Nε-acetylated proteins | [53] |
| Root/hypocotyl | Soybean | QProteome Mitochondrial Isolation kit | 2DE, LC–MS/MS | 327 proteins | [25] |
| Endoplasmic reticulum | |||||
| Root tip | Soybean | Endoplasmic Reticulum Enrichment kit | LC–MS/MS | 255, 368, 103 proteins in control, flooding, drought | [11] |
| Cell wall | |||||
| Cell culture | Sugarcane | Washings of cell walls with 5 mM acetate buffer | 1DE, LC–MS/MS | 377 proteins | [57] |
| Mature stem | Arabidopsis | Washings of cell walls with 5 mM acetate buffer | 1DE, LC–MS/MS | 302 cell wall proteins | [58] |
| Rosettes | Arabidopsis | Washings of cell walls with 5 mM acetate buffer | 1DE, LC–MS/MS | 361 cell wall proteins | [59] |
| Plasma membrane | |||||
| Seedlings | Arabidopsis | Density gradient, Free-flow electrophoresis | LC–MS/MS | 1029 proteins | [60] |
| Seedlings | Arabidopsis | Density gradient | 2D-DIGE, LC-MS/MS, MALDI-TOF/MS | 36 microdomain proteins | [61] |
| Seedlings | Oat, Rye | Density gradient | LC–MS/MS | 740, 809 proteins in oat, rye | [62] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komatsu, S.; Hashiguchi, A. Subcellular Proteomics: Application to Elucidation of Flooding-Response Mechanisms in Soybean. Proteomes 2018, 6, 13. https://doi.org/10.3390/proteomes6010013
Komatsu S, Hashiguchi A. Subcellular Proteomics: Application to Elucidation of Flooding-Response Mechanisms in Soybean. Proteomes. 2018; 6(1):13. https://doi.org/10.3390/proteomes6010013
Chicago/Turabian StyleKomatsu, Setsuko, and Akiko Hashiguchi. 2018. "Subcellular Proteomics: Application to Elucidation of Flooding-Response Mechanisms in Soybean" Proteomes 6, no. 1: 13. https://doi.org/10.3390/proteomes6010013




