Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets
Abstract
:1. Introduction
2. Systemic Treatment of Gastric Carcinoma
2.1. Chemotherapy
2.2. Targeted Therapy
2.2.1. Mitogenic Signaling Pathways as Therapeutic Targets
2.2.2. Signaling Pathways in Angiogenesis as Therapeutic Targets
2.2.3. Immune Checkpoint Molecules as Targets for Therapy
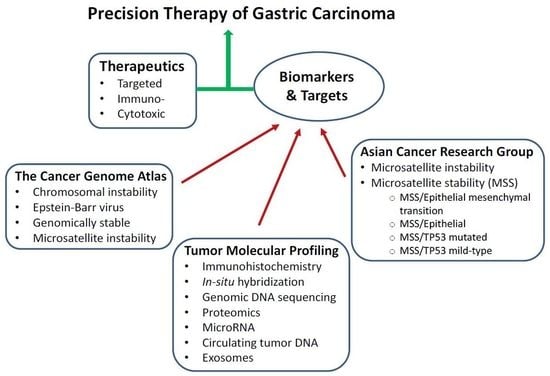
3. Molecular Classification and Profiling of Gastric Carcinoma
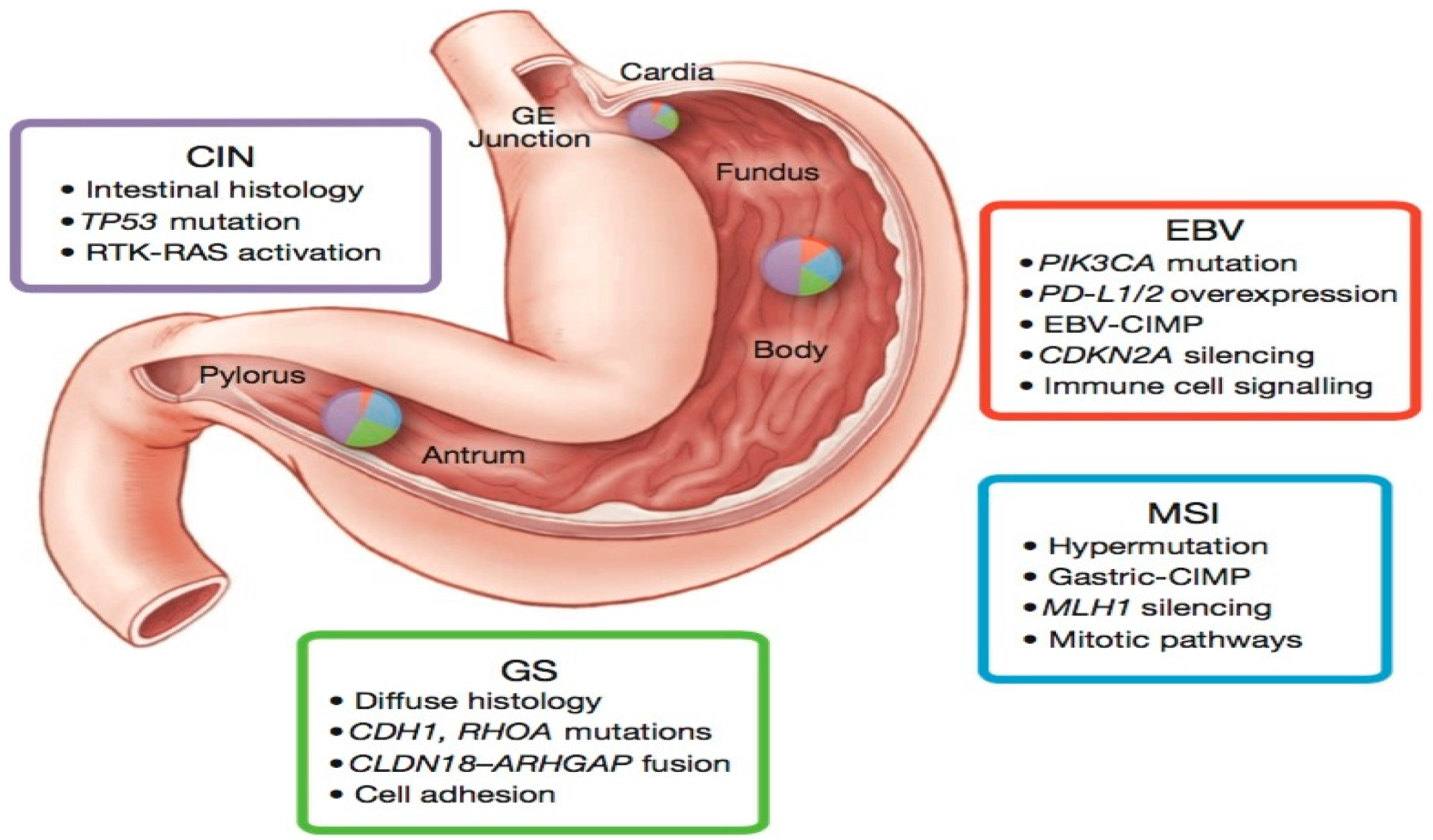
3.1. TCGA Sub-Typing of Gastric Carcinoma: Potential Therapeutic Targets
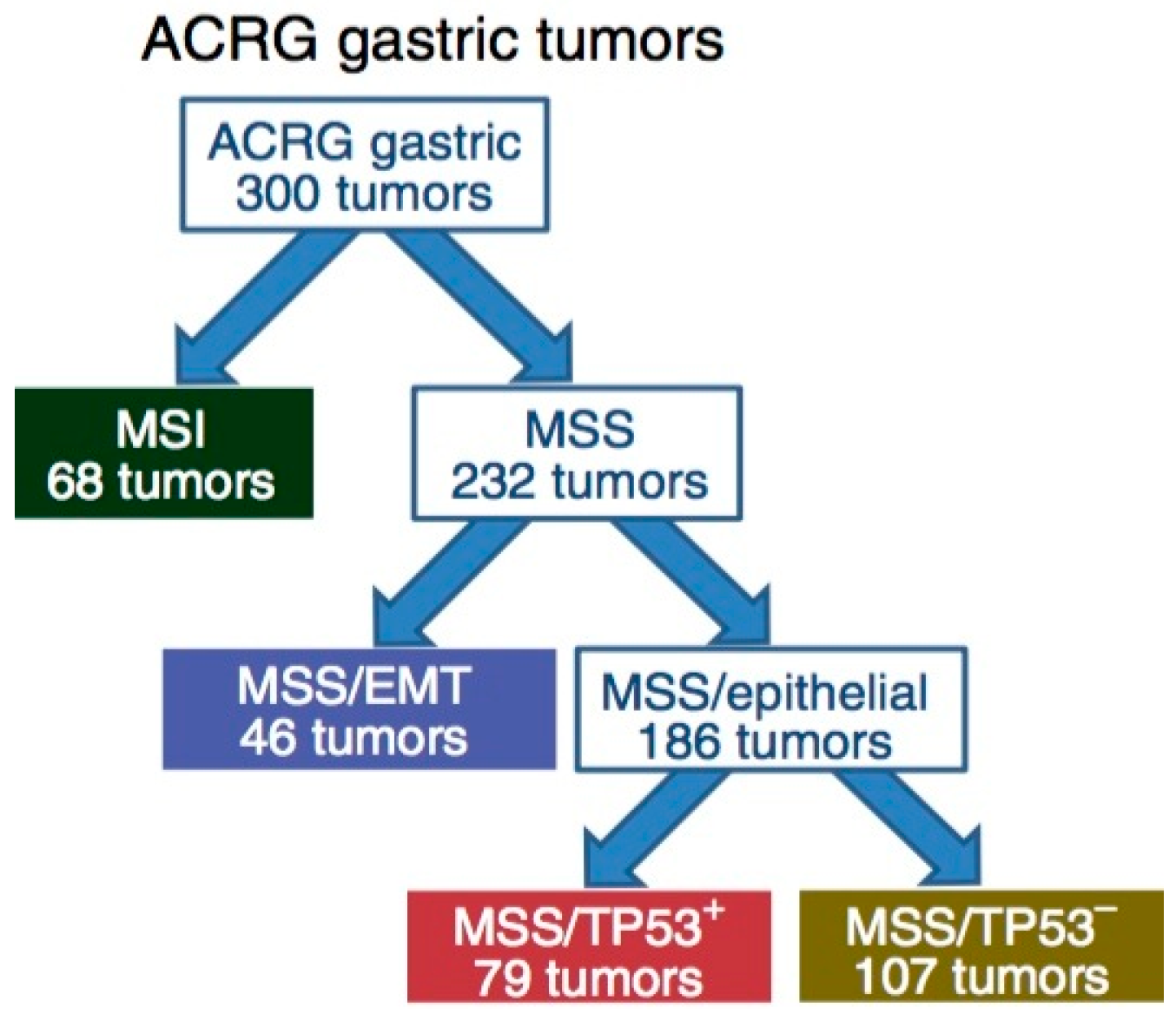
3.2. ACRG Sub-Typing of Gastric Carcinoma: Potential Prognostic Biomarkers
3.3. Comparison of TCGA and ACRG Data
4. Molecular Profiling of Gastric Carcinoma: Therapeutic Targets and Predictive Biomarkers
4.1. Therapeutic Targets
4.2. Predictive Biomarkers
5. Conclusions and Perspectives
Author Contributions
Conflicts of Interest
References
- International Agency for Research on Cancer. Stomach Cancer. Estimated Incidence, Mortality and Prevalence Worldwide in 2012. Available online: http://globocan.iarc.fr/old/FactSheets/cancers/stomach-new.asp (accessed on 28 October 2017).
- Colvin, H.; Yamamoto, K.; Wada, N.; Mori, M. Hereditary gastric cancer syndromes. Surg. Oncol. Clin. N. Am. 2015, 24, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Lunet, N.; Valbuena, C.; Virira, A.; Lopes, C.; Lopes, C.; David, L.; Careneiro, F.; Barros, H. Fruit and vegetable consumption and gastric cancer by location and histological type: Case-control and meta-analysis. Eur. J. Cancer Prev. 2007, 16, 312–327. [Google Scholar] [CrossRef] [PubMed]
- Ladeiras-Lopes, R.; Pereira, A.; Nogueira, A.; Pinheiro-Torres, T.; Pinto, I.; Santos-Pereira, R.; Lunet, N. Smoking and gastric cancer: Systemic review and meta-analysis of cohort studies. Cancer Causes Control. 2008, 19, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhou, Y.; Chen, B.; Wan, H.W.; Jia, G.Q.; Bai, H.L.; Wu, X.T. Overweight, obesity and gastric cancer risk: Results from a meta-analysis of cohort studies. Eur. J. Cancer 2009, 45, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Pinheiro, H.; Figueiredo, J.; Seruca, R.; Carneiro, F. Familial gastric cancer: Genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015, 16, e60–e70. [Google Scholar] [CrossRef]
- Crew, K.D.; Neugut, A.I. Epidemiology of gastric cancer. World J. Gastroenterol. 2006, 12, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Wu, M.S.; Shun, C.T.; Wang, H.P.; Lin, C.C.; Lin, J.T. Lymphoepithelioma-like carcinoma of the stomach: A subset of gastric carcinoma with distinct clinicopathological features and high prevalence of Epstein-Barr virus infection. Hepatogastroenterology 1999, 46, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.S.; Shun, C.T.; Wu, C.C.; Hsu, T.Y.; Lin, M.T.; Chang, M.C.; Wang, H.P.; Lin, J.T. Epstein-Barr virus-associated gastric carcinomas: Relation to H. pylori infection and genetic alterations. Gastroenterology 2000, 118, 1031–1038. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. IARC Working Group Reports. Helicobacter Pylori: Eradication as A Strategy for Preventing Gastric Cancer. Available online: http://www.iarc.fr/ en/publications /pdfs-online/wrk/wrk8 (accessed on 28 October 2017).
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 1302. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Doi, T.; Woo-Jun Jang, R.; Muro, K.; Satoh, T.; Machado, M. KEYNOTE-059 cohort 1: Efficacy and safety of pembrolizumab (pembro) monotherapy in patients with previously treated advanced gastric cancer. J. Clin. Oncol. 2017, 35 (Suppl. 15), 4003. [Google Scholar] [CrossRef]
- Kankeu Fonkoua, L.A.; Liao, J.; Yee, N.S. Molecular profiling-guided therapy in gastroesophageal carcinoma: A single-institution experience. J. Clin. Oncol. 2017, 35 (Suppl. 15), e15582. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Bosman, F.T.; Carneiro, F.; Hruban, R.H.; Thiese, N.D. WHO Classification of Tumors of the Digestive System, 4th ed.; IARC: Lyon, France, 2010; pp. 44–58. [Google Scholar]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Rivera, F.; Vega-Villegas, M.E.; Lopez-Brea, M.F. Chemotherapy of advanced gastric cancer. Cancer Treat. Rev. 2007, 33, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, R.; Lee, C.; Kim, R. Is there a role for second-line chemotherapy in advanced gastric cancer? Lancet Oncol. 2009, 10, 903–912. [Google Scholar] [CrossRef]
- Okines, A.F.; de Castro, D.G.; Cunningham, D.; Chau, I.; Langley, R.E.; Thompson, L.C.; Stenning, S.P.; Saffery, C.; Barbachano, Y.; Coxon, F. Biomarker analysis in oesophagogastric cancer: Results from the REAL3 and TransMAGIC trials. Eur. J. Cancer 2013, 49, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Moiseyenko, V.; Tjulandin, S.; Majlis, A.; Constenla, M.; Boni, C.; Rodrigues, A.; Fodor, M.; Chao, Y.; Voznyi, E. Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: A report of the V325 Study Group. J. Clin. Oncol. 2006, 24, 4991–4997. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; Starling, N.; Rao, S.; Iveson, T.; Nicolson, M.; Coxon, F.; Middleton, G.; Daniel, F.; Oates, J.; Norman, A.R. Capecitabine and oxaliplatin for advanced esophagogastric cancer. N. Engl. J. Med. 2008, 358, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Al-Batran, S.; Hartmann, J.; Probst, S.; Schmalenberg, H.; Hollerbach, S.; Hofheinz, R.; Rethwisch, V.; Seipelt, G.; Homann, N.; Wilhelm, G. Phase III trial in metastatic gastroesophageal adenocarcinoma with fluorouracil, leucovorin plus either oxaliplatin or cisplatin: A study of the Arbeitsgemeinschaft Internistische Onkologie. J. Clin. Oncol. 2008, 26, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.; Rodriguez, W.; Bodoky, G.; Moiseyenko, V.; Lichinitser, M.; Gorbunova, V.; Vynnychenko, I.; Garin, A.; Lang, I.; Falcon, S. Multicenter phase III comparison of cisplatin/S-1 with cisplatin/infusional fluorouracil in advanced gastric or gastroesophageal adenocarcinoma study: The FLAGS trial. J. Clin. Oncol. 2010, 28, 1547–1553. [Google Scholar] [CrossRef] [PubMed]
- Hudis, C.A. Trastuzumab—Mechanism of action and use in clinical practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Han, S.; Ding, S.; He, J.; Zhang, H. Antibody-nanoparticle conjugate constructed with trastuzumab and nanoparticle albumin-bound paclitaxel for targeted therapy of human epidermal growth factor receptor 2-positive gastric cancer. Oncol. Rep. 2018, 39, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Kowanetz, M.; Ferrara, N. Vascular endothelial growth factor signaling pathways: Therapeutic perspective. Clin. Cancer Res. 2006, 12, 5018–5022. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Iljin, K.; Dumont, D.J.; Alitalo, K. Tie receptors: New modulators of angiogenic and lymphangiogenic responses. Nat. Rev. Mol. Cell Biol. 2001, 2, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Carbone, C.; Piro, G.; Merz, V.; Simionato, F.; Santoro, R.; Zecchetto, C.; Tortora, G.; Melisi, D. Angiopoietin-like proteins in angiogenesis, inflammation and cancer. Int. J. Mol. Sci. 2018, 19, 431. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.-H.; Lennartsson, J.; Westermark, B. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J. Int. Med. 2018, 283, 16–44. [Google Scholar] [CrossRef] [PubMed]
- Ronca, R.; Giacomini, A.; Rusnati, M.; Presta, M. The potential of fibroblast growth factor/fibroblast growth factor receptor signaling as a therapeutic target in tumor angiogenesis. Expert Opin. Ther. Targets 2015, 19, 1361–1377. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.M.; Wang, F.; Zheng, Y.; Fu, Z.Z.; Zheng, L.; Chen, L.L. Roles of fibroblast activation protein and hepatocyte growth factor expressions in angiogenesis and metastasis of gastric cancer. Pathol. Oncol. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.J.; Other-Gee Pohl, S.; Deshmukh, A.; Visweswaran, M.; Ward, N.C.; Arfuso, F.; Agostino, M.; Dharmarajan, A. The role of Wnt signaling in angiogenesis. Clin. Biochem. Rev. 2017, 38, 131–142. [Google Scholar] [PubMed]
- Tanaka, T.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitsui, A.; Katada, T.; Shiozaki, M.; Naganawa, Y.; Fujii, Y.; Takeyama, H. Vascular endothelial growth factor C (VEGF-C) in esophageal cancer correlates with lymph node metastasis and poor patient prognosis. J. Exp. Clin. Cancer Res. 2010, 29, 83. [Google Scholar] [CrossRef] [PubMed]
- Omoto, I.; Matsumoto, M.; Okumura, H.; Uchikado, Y.; Setoyama, T.; Kita, Y.; Owaki, T.; Kijima, Y.; Shinchi, H.; Ishigami, S. Expression of vascular endothelial growth factor-C and vascular endothelial growth factor receptor-3 in esophageal squamous cell carcinoma. Oncol. Lett. 2014, 7, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; Vieira dos Santos, L.; Aprile, G.; Ferry, D. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Wilke, H.; Muro, K.; Van Cutsem, E.; Oh, S.C.; Bodoky, G.; Shimada, Y.; Hironaka, S.; Sugimoto, N.; Lipatov, O.; Kim, T.Y. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-esophageal junction adenocarcinoma (RAINBOW): A double-blind, randomised phase 3 trial. Lancet Oncol. 2014, 15, 1224–1235. [Google Scholar] [CrossRef]
- Gu, L.; Chen, M.; Guo, D.; Zhu, H.; Zhang, W.; Pan, J.; Zhong, X.; Li, X.; Qian, H.; Wang, X. PD-L1 and gastric cancer prognosis: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0182692. [Google Scholar] [CrossRef] [PubMed]
- Muro, K.; Bang, Y.; Shankaran, V.; Geva, R.; Catenacci, D.V.T.; Gupta, S.; Eder, J.P.; Berger, R.; Gonzalez, E.J.; Pulini, J. A phase 1B study of pembrolizumab (PEMBRO; MK-3475) in patients with advanced gastric cancer. Ann. Oncol. 2014, 25, 1–41. [Google Scholar] [CrossRef]
- Muro, K.; Chung, H.C.; Shankaran, V.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Golan, T.; Le, D.T.; Burtness, B.; et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): A multicenter, open-label, phase 1b trial. Lancet Oncol. 2016, 17, 717–726. [Google Scholar] [CrossRef]
- Miura, J.T.; Xiu, J.; Thomas, J.; George, B.; Carron, B.R.; Tsai, S.; Johnston, F.M.; Turaga, K.K.; Gamblin, T.G. Tumor profiling of gastric and esophageal carcinoma reveal different treatment options. Cancer Biol. Ther. 2015, 16, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Deng, N.; Goh, L.K.; Wang, H.; Das, K.; Tao, J.; Tan, I.B.; Zhang, S.; Lee, M.; Wu, J.; Lim, K.H. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 2012, 61, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.M.; Sanford, E.M.; Klempner, S.J.; Rubinson, D.A.; Wang, K.; Palma, N.A.; Chmielecki, J.; Yelensky, R.; Palmer, G.A.; Morosini, D. Prospective comprehensive genomic profiling of advanced gastric carcinoma cases reveals frequent clinically relevant genomic alterations and new routes for targeted therapies. Oncologist 2015, 20, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 373, 1979. [Google Scholar] [CrossRef]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migration: Integrating signals from front to back. Science 2003, 302, 1704–1709. [Google Scholar] [CrossRef] [PubMed]
- Thumkeo, D.; Watanabe, S.; Narumiya, S. Physiological roles of Rho and Rho effectors in mammals. Eur. J. Cell. Biol. 2013, 92, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I. Principal Component Analysis; Wiley Online Library: New York, NY, USA, 2002. [Google Scholar]
- Loboda, A.; Nebozhyn, M.V.; Watters, J.W.; Buser, C.A.; Shaw, P.M.; Huang, P.S.; Van’t Veer, L.; Tollenar, R.A.E.M.; Jackson, D.B.; Agrawal, D. EMT is the dominant program in human colon cancer. BMC Med. Genom. 2011, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Coppola, D.; Nebozhyn, M.; Khalil, F.; Dai, H.; Yeatman, T.; Loboda, A.; Mule, J.J. Unique ectopic lymph node-like structures present in human primary colorectal carcinoma are identified by immune gene array profiling. Am. J. Pathol. 2011, 179, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Van’t Veer, L.; Lamb, J.; He, Y.D.; Mao, M.; Fine, B.M.; Bernards, R.; Van de Vijver, M.; Deutsch, P.; Sachs, A. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005, 65, 4059–4066. [Google Scholar] [CrossRef] [PubMed]
- Zouridis, H.; Deng, N.; Ivanova, T.; Zhu, Y.; Wong, B.; Huang, D.; Wu, Y.H.; Wu, Y.; Tan, I.B.; Liem, N.; et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci. Transl. Med. 2012, 4, 156ra140. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Benita, Y.; Cao, Z.; Giallourakis, C.; Li, C.; Gardet, A.; Xavier, R.J. Gene enrichment profiles reveal T cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 2010, 115, 5376–5384. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Sato, F.; Selaru, F.M.; Olaru, A.; Perry, K.; Kimos, M.C.; Tamura, G.; Matsubara, N.; Wang, S.; Xu, Y.; et al. Instabilotyping reveals unique mutational spectra in microsatellite unstable gastric cancers. Cancer Res. 2002, 62, 3641–3645. [Google Scholar] [PubMed]
- Mori, Y.; Selaru, F.M.; Sato, F.; Yin, J.; Simms, L.; Xu, Y.; Olaru, A.; Deacu, E.; Wang, S.; Taylor, J.M.; et al. The impact of microsatellite instability on the molecular phenotype of colorectal tumors. Cancer Res. 2003, 63, 4577–4582. [Google Scholar] [PubMed]
- Christman, K.; Kelsen, D.; Saltz, L.; Tarassoff, P.G. Phase II trial of gemcitabine in patients with advanced gastric cancer. Cancer 1994, 73, 5–7. [Google Scholar] [CrossRef]
- De Lange, S.M.; van Groeningen, C.J.; Kroep, J.R.; Van Bochove, A.; Snijders, J.F.; Peters, G.J. Phase II trial of cisplatin and gemcitabine in patients with advanced gastric cancer. Ann. Oncol. 2004, 15, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Hao, Y.; Wei, Y.; Yin, Q.; Du, J.; Zhao, X. Response to first-line chemotherapy in patients with non-small cell lung cancer according to RRM1 expression. PLoS ONE 2014, 9, e92320. [Google Scholar] [CrossRef] [PubMed]
- Yee, N.S.; Fonkoua, L.A.K.; Liao, J. Molecular Profiling of Gastro-Esophageal Carcinoma as Basis for Precision Cancer Medicine. In Proceedings of the BIT’s 8th World Gene Conference—2017, Macao, China, 15 November 2017. [Google Scholar]
- Aichler, M.; Luber, B.; Lordick, F.; Walch, A. Proteomic and metabolic prediction of response to therapy in gastric cancer. World J. Gastroenterol. 2014, 14, 13648–13657. [Google Scholar] [CrossRef] [PubMed]
- Balluff, B.; Frese, C.; Maier, S.K.; Schone, C.; Kuster, B.; Schmitt, M.; Aubele, M.; Hofler, H.; Deelder, A.M.; Heck, A.J.R.; et al. De novo discovery of phenotypic intratumor heterogeneity using imaging mass spectrometry. J. Pathol. 2015, 235, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Piga, I.; Galli, M.; Stella, M.; Denti, V.; Del Puppo, M.; Magni, F. Matrix-assisted laser desorption/ionization mass spectrometry imaging in the study of gastric cancer: A mini review. Int. J. Mol. Sci. 2017, 18, 2588. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Hsu, S.-D.; Huang, W.-Y.; Huang, H.-Y.; Chen, W.L.; Weng, S.-L.; Huang, H.D. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014, 3, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, K.; Xi, H.; Cai, A.; Wu, X.; Cui, J.; Li, J.; Qiao, Z.; Wei, B.; Chen, L. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: A meta-analysis. Oncotarget 2017, 8, 6330–6340. [Google Scholar] [CrossRef] [PubMed]
- Biankin, A.V.; Piantadosi, S.; Hollingsworth, S.J. Patient-centric trials for therapeutic development in precision oncology. Nature 2015, 526, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Catenacci, D.V.T.; Polite, B.N.; Henderson, L.; Xu, P.; Rambo, B.; Liao, W.L.; Hembrough, T.A.; Zhao, L.; Xiao, S.Y.; Hart, J.; et al. Towards personalized treatment for gastroesophageal adenocarcinoma (GEC): Strategies to address tumor heterogeneity. J. Clin. Oncol. 2014, 32, 66. [Google Scholar] [CrossRef]
| Treatment | Patients (n) | RR (%) | PFS (months) | OS (months) | Reference |
|---|---|---|---|---|---|
| CF vs. DCF | 224 vs. 221 | 25 vs. 37 | 3.7 vs. 5.6 (* p < 0.001) | 8.6 vs. 9.2 (* p < 0.02) | [22] |
| ECF vs. ECX vs. EOF vs. EOX | 263 vs. 250 vs. 245 vs. 244 | 38 vs. 41 vs. 40 vs. 47 | 6.2 vs. 6.7 vs. 6.5 vs. 7.0 (NS) | 9.9 vs. 9.9 vs. 9.3 vs. 11.2 (NS) | [23] |
| 5-FU + LV + cisplatin vs. 5-FU + LV + oxaliplatin | 112 vs. 106 | 25 vs. 34 | 3.9 vs. 5.8 (NS) | 8.8 vs. 10.7 (NS) | [24] |
| Cisplatin + 5-FU vs. Cisplatin + S-1 | 508 vs. 521 | 31 vs. 29 | 5.6 vs. 5.3 (NS) | 7.9 vs. 8.6 (NS) | [25] |
| Subtypes | Targets | Targeted Agents |
|---|---|---|
| EBV | PIK3CA | Idelalisib, Taselisib |
| PD-L1/L2 | Pembrolizumab, Nivolumab, Durvalumab, Avelumab, Atezolizumab | |
| MSI | MLH1 silencing | Pembrolizumab, Nivolumab, Durvalumab, Avelumab, Atezolizumab |
| PIK3CA | Idelalisib, Taselisib | |
| EGFR | Erlotinib, Gefitinib | |
| ERBB2 | Trastuzumab | |
| ERBB3 | Pertuzumab | |
| PD-L1 | Pembrolizumab, Nivolumab, Durvalumab, Avelumab, Atezolizumab | |
| CIN | EGFR | Erlotinib, Gefitinib |
| VEGFA | Bevacizumab, Ramucirumab | |
| CCNE1, CCND1, CDK6 | Palbociclib, Ribociclib, Abemaciclib | |
| GS | RHOA | - |
| CLDN18 | - |
| Genetic Alteration | TCGA | ACRG | ||||||
|---|---|---|---|---|---|---|---|---|
| MSI | EBV | GS | CIN | MSI | MSS/EMT | MSS/TP53+ | MSS/TP53− | |
| HER2 amp | 0 | 12 | 3 | 22 | 0 | 0 | 3.0 | 17.4 |
| HER2 mut | 11 | 4 | 3 | 3 | 16.3 | 2.8 | 0 | 4.7 |
| MET amp | 2 | 0 | 0 | 7 | 1.6 | 0 | 3.0 | 3.5 |
| PIK3CA amp | 3 | 8 | 2 | 7 | 0 | 0 | 0 | 1.1 |
| PIK3CA mut | 42 | 77 | 10 | 3 | 32.6 | 8.3 | 16.9 | 4.7 |
| KRAS mut | 23 | 4 | 9 | 5 | 23.3 | 0 | 8.5 | 3.5 |
| RHOA mut | 5 | 8 | 14 | 2 | 0 | 2.8 | 6.8 | 3.5 |
| CDH1 mut | 8 | 0 | 34 | 3 | 7.0 | 2.8 | 1.7 | 3.5 |
| FGFR2 amp | 0 | 0 | 7 | 7 | 0 | 4.9 | 3.0 | 1.2 |
| BRAF mut | 22 | 8 | 0 | 0 | 11.6 | 2.8 | 1.7 | 3.5 |
| ALK mut | 9 | 0 | 5 | 2 | 16.3 | 0 | 0 | 2.4 |
| ARID1A mut | 84 | 54 | 16 | 9 | 44.2 | 13.9 | 18.6 | 5.9 |
| TP53 mut | 39 | 4 | 14 | 70 | 25.6 | 33.3 | 23.7 | 60 |
| PTEN mut | 25 | 15 | 2 | 1 | 14 | 5.6 | 3.4 | 3.5 |
| MTOR mut | 30 | 4 | 3 | 1 | 14 | 0 | 1.7 | 3.5 |
| APC mut | 36 | 0 | 3 | 12 | 16.3 | 2.8 | 15.3 | 8.2 |
| FBXW7 mut | 34 | 0 | 5 | 1 | 16.3 | 2.8 | 1.7 | 2.4 |
| SMAD4 mut | 8 | 12 | 9 | 7 | 4.7 | 2.8 | 8.5 | 2.4 |
| Biomarker | Number of Specimens (%) | Beneficial Agents |
|---|---|---|
| TS (−) | 19 (70.4) | Fluorouracil, Capecitabine |
| TOPO1 (+) * | 16 (59.3) | Irinotecan, Topotecan |
| PTEN (−) | 11 (40.7) | Trastuzumab, anti-EGFR |
| ERCC1 (−) * | 11 (40.7) | Cisplatin, Carboplatin, Oxaliplatin |
| TOP2A (+) * | 11 (40.7) | Doxorubicin, Epirubicin |
| RRM1 (−) | 10 (37.0) | Gemcitabine |
| MGMT (−) | 9 (33.3) | Temozolomide, Dacarbazine |
| TUBB3 (−) | 8 (29.6) | Docetaxel, nab-paclitaxel, paclitaxel |
| cMET (+) | 7 (25.9) | Anti-MET |
| TLE3 (+) | 6 (22.2) | Docetaxel, Paclitaxel |
| SPARC Mono (+) | 5 (18.5) | nab-Paclitaxel |
| SPARC Poly (+) | 4 (14.8) | nab-Paclitaxel |
| HER2 (+) * | 4 (14.8) | Trastuzumab, Lapatinib |
| PGP (−) * | 4 (14.8) | Taxane |
| Biomarker | Method | Beneficial Agent | Treatment Prior to MP | MP-Based Treatment | PFS Ratio |
|---|---|---|---|---|---|
| HER2/Neu amplification | FISH, IHC | Trastuzumab | Docetaxel + Irinotecan PFS = 2.2 months | Trastuzumab + Docetaxel + Irinotecan PFS = 6.3 months | 2.9 |
| Topoisomerase 1 positive | IHC | Irinotecan, Topotecan | Epirubicin + Oxaliplatin + Capecitabine PFS = 2.3 months | Docetaxel + Irinotecan PFS = 4.5 months | 2.0 |
| SPARC Monoclonal positive | IHC | nab-Paclitaxel | Docetaxel + Irinotecan PFS = 1.9 months | Gemcitabine + nab-Paclitaxel PFS = 3.6 month | 1.9 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kankeu Fonkoua, L.; Yee, N.S. Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets. Biomedicines 2018, 6, 32. https://doi.org/10.3390/biomedicines6010032
Kankeu Fonkoua L, Yee NS. Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets. Biomedicines. 2018; 6(1):32. https://doi.org/10.3390/biomedicines6010032
Chicago/Turabian StyleKankeu Fonkoua, Lionel, and Nelson S. Yee. 2018. "Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets" Biomedicines 6, no. 1: 32. https://doi.org/10.3390/biomedicines6010032