Targeted Stimulation Using Differences in Activation Probability across the Strength–Duration Space
Abstract
:1. Introduction
2. Methods
2.1. Cortical Cell Culture
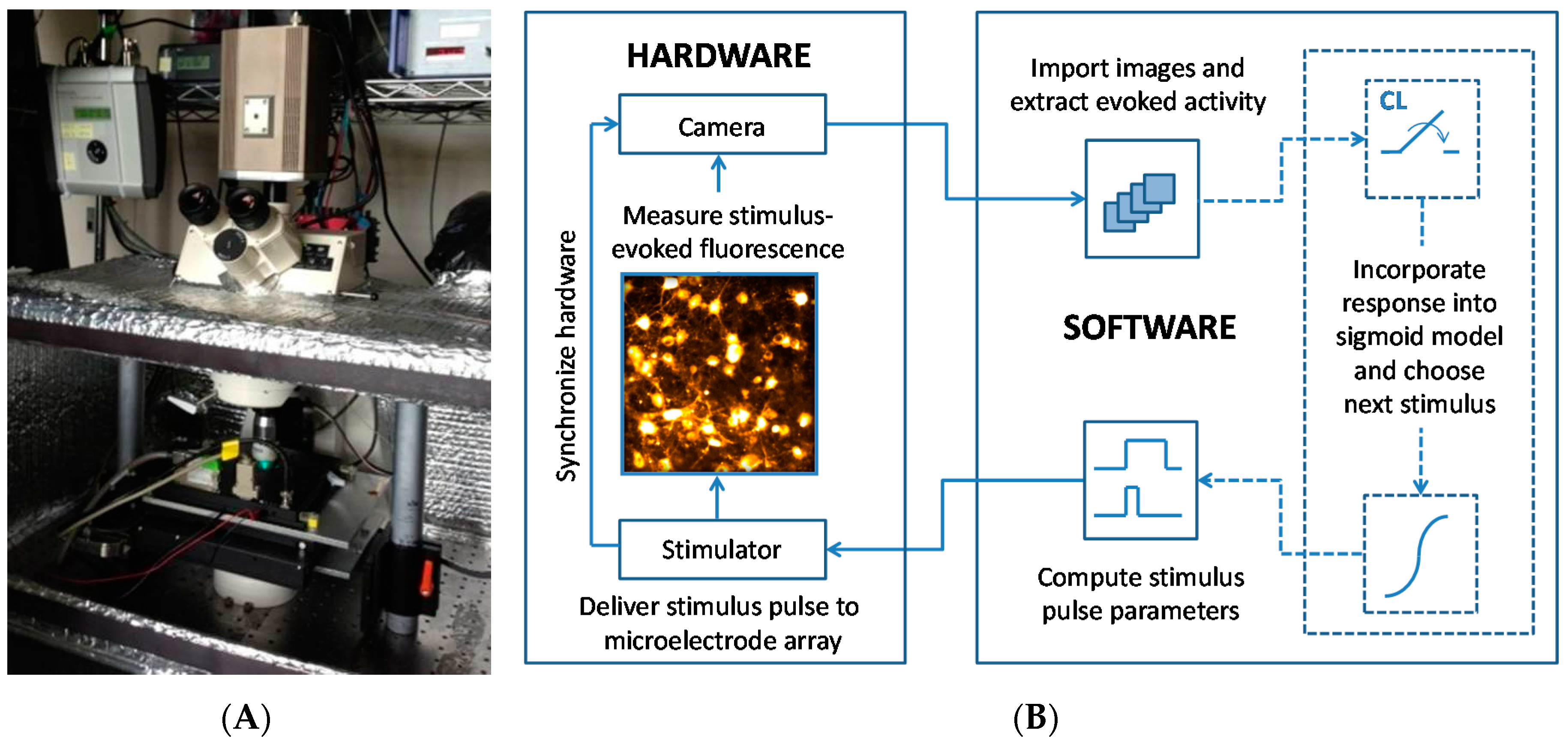
2.2. Electrical Stimulation
2.3. Optical Imaging
2.4. The Closed-Loop Search Algorithm and Activation Models
3. Results
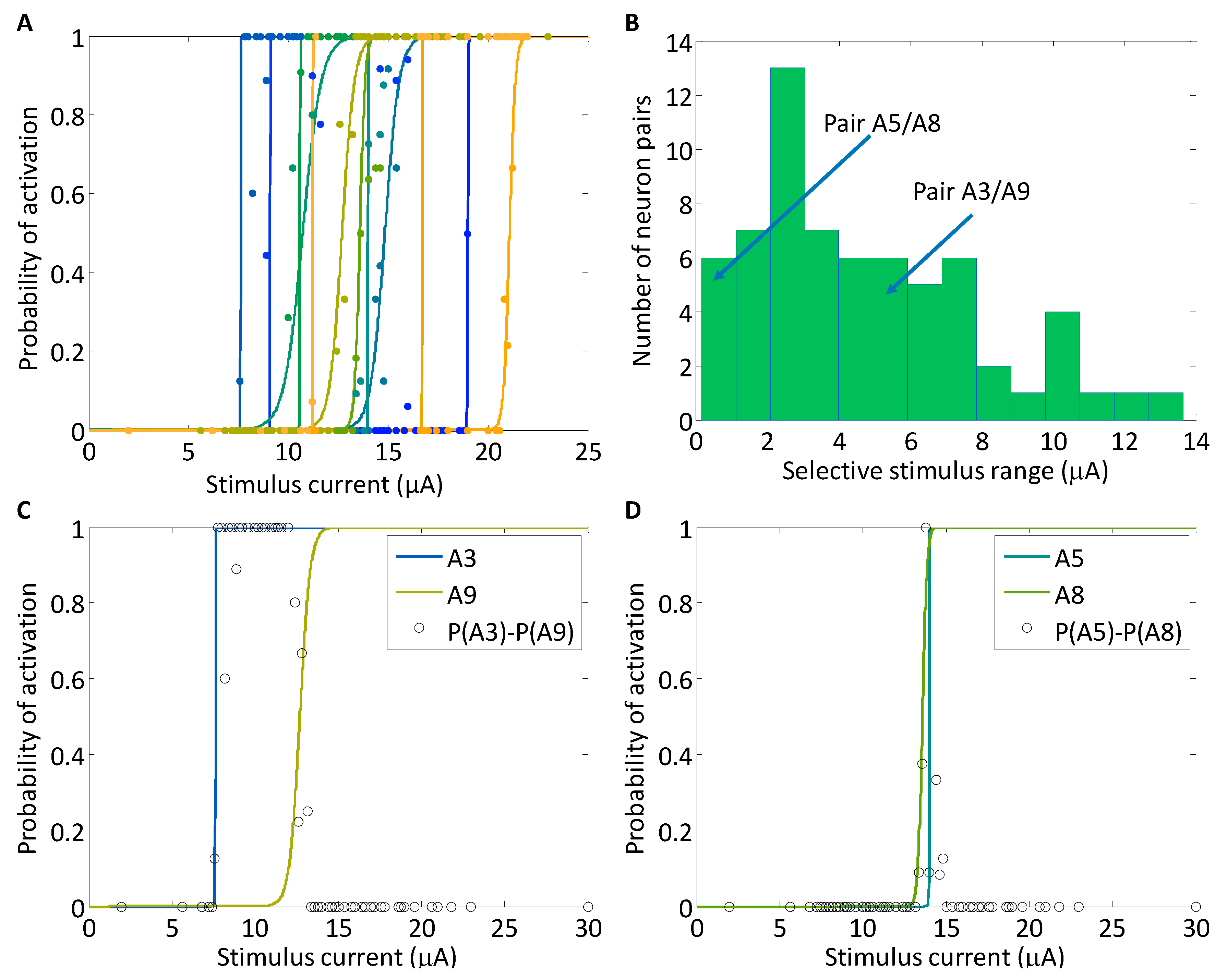
3.1. Analysis of Selectivity with a One-Dimensional Input Space
3.2. Analysis of Selectivity with a Two-Dimensional Input Space
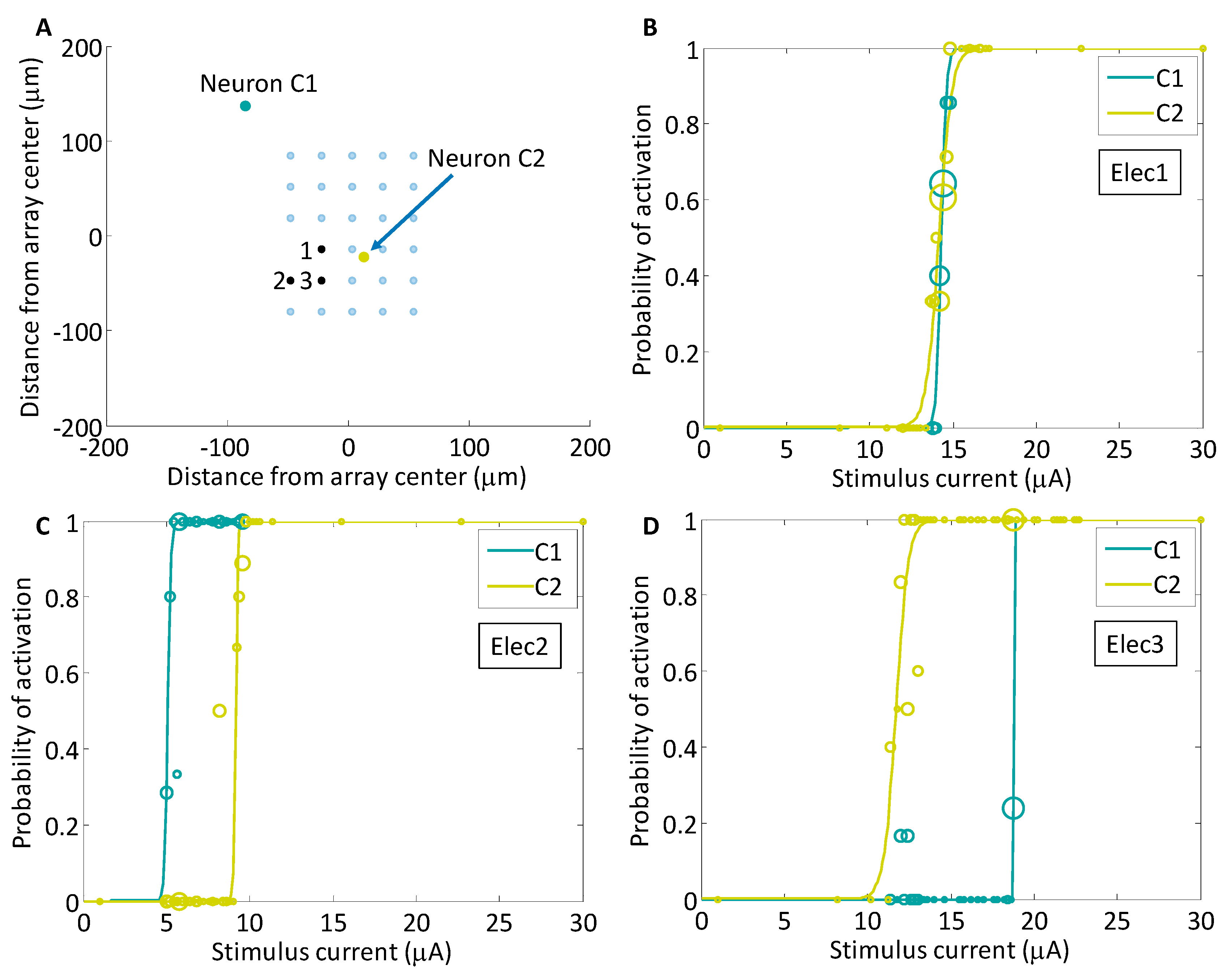
3.3. Analysis of Selectivity with Multiple Electrodes
4. Discussion
4.1. Adaptive Targeted Searches Can Find Even a Small Window for Selectivity
4.2. Probing of the Population Response Is Essential for Targeted Stimulation
4.3. Using an Array of Spatially Distributed Electrodes
4.4. As the Parameter Space Increases, CL Systems Behave More Similarly to OL Systems
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clark, K.L.; Armstrong, K.M.; Moore, T. Probing neural circuitry and function with electrical microstimulation. Proc. R. Soc. B Biol. Sci. 2011, 278, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Borchers, S.; Himmelbach, M.; Logothetis, N.; Karnath, H.O. Direct electrical stimulation of human cortex—The gold standard for mapping brain functions? Nat. Rev. Neurosci. 2011, 13, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Joucla, S.; Branchereau, P.; Cattaert, D.; Yvert, B. Extracellular neural microstimulation may activate much larger regions than expected by simulations: A combined experimental and modeling study. PLoS ONE 2012, 7, e41324. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.B.; Smith, K.L.; Shain, W.; Dowell-Mesfin, N.M.; Kim, S.J.; Hynd, M.R. Optical monitoring of neural networks evoked by focal electrical stimulation on microelectrode arrays using FM dyes. Med. Biol. Eng. Comput. 2010, 48, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.P.; Zeller-Townson, R.; Fong, M.F.; Desai, S.A.; Gross, R.E.; Potter, S.M. Closed-loop, multichannel experimentation using the open-source NeuroRighter electrophysiology platform. Front. Neural Circuits 2013, 6, 98. [Google Scholar] [CrossRef] [PubMed]
- Franke, F.; Jäckel, D.; Dragas, J.; Müller, J.; Radivojevic, M.; Bakkum, D.; Hierlemann, A. High-density microelectrode array recordings and real-time spike sorting for closed-loop experiments: An emerging technology to study neural plasticity. Front. Neural Circuits 2012, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Bakkum, D.J.; Hierlemann, A. Sub-millisecond closed-loop feedback stimulation between arbitrary sets of individual neurons. Front. Neural Circuits 2012, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, J.; Hendriksen, P.J.; van Kleef, R.G.; de Groot, A.; Bovee, T.F.; Rietjens, I.M.; Westerink, R.H. Detection of marine neurotoxins in food safety testing using a multielectrode array. Mol. Nutr. Food Res. 2014, 58, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, P.; Martin, M.; LeFew, W.R.; Ross, J.; Houck, K.A.; Shafer, T.J. Multi-well microelectrode array recordings detect neuroactivity of ToxCast compounds. Neurotoxicology 2014, 44, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, E.; Roberts, W.J. Synaptic actions of single interneurones mediating reciprocal Ia inhibition of motoneurones. J. Physiol. 1972, 222, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.J.; Ziv, O.R.; Rizzo, J.F., III; Scribner, D.; Johnson, L. Spatiotemporal aspects of pulsed electrical stimuli on the responses of rabbit retinal ganglion cells. Exp. Eye Res. 2009, 89, 972–979. [Google Scholar] [CrossRef] [PubMed]
- Sekirnjak, C.; Hottowy, P.; Sher, A.; Dabrowski, W.; Litke, A.M.; Chichilnisky, E.J. Electrical stimulation of mammalian retinal ganglion cells with multielectrode arrays. J. Neurophysiol. 2006, 95, 3311–3327. [Google Scholar] [CrossRef] [PubMed]
- Yeomans, J.S.; Maidment, N.T.; Bunney, B.S. Excitability properties of medial forebrain bundle axons of A9 and A10 dopamine cells. Brain Res. 1988, 450, 86–93. [Google Scholar] [CrossRef]
- Bostock, H. The strength-duration relationship for excitation of myelinated nerve: Computed dependence on membrane parameters. J. Physiol. 1983, 341, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Nowak, L.G.; Bullier, J. Axons, but not cell bodies, are activated by electrical stimulation in cortical gray matter. Exp. Brain Res. 1998, 118, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Dziubińska, H.; Paszewski, A.; Trębacz, K.; Zawadzki, T. Electrical activity of the liverwort Conocephalum conicum: The all-or-nothing law, strength-duration relation, refractory periods and intracellular potentials. Physiol. Plant. 1983, 57, 279–284. [Google Scholar] [CrossRef]
- Fozzard, H.A.; Schoenberg, M. Strength-duration curves in cardiac Purkinje fibres: Effects of liminal length and charge distribution. J. Physiol. 1972, 226, 593–618. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, C.C.; Grill, W.M. Extracellular stimulation of central neurons: Influence of stimulus waveform and frequency on neuronal output. J. Neurophysiol. 2002, 88, 1592–1604. [Google Scholar] [PubMed]
- Boinagrov, D.; Loudin, J.; Palanker, D. Strength–duration relationship for extracellular neural stimulation: Numerical and analytical models. J. Neurophysiol. 2010, 104, 2236–2248. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, A.; Amodaj, N.; Hoover, K.; Vale, R.; Stuurman, N. Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol. 2010, 14, 14–20. [Google Scholar]
- Potter, S.M.; DeMarse, T.B. A new approach to neural cell culture for long-term studies. J. Neurosci. Methods 2001, 110, 17–24. [Google Scholar] [CrossRef]
- Wagenaar, D.A.; Pine, J.; Potter, S.M. Effective parameters for stimulation of dissociated cultures using multi-electrode arrays. J. Neurosci. Methods 2004, 138, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Bakkum, D.J.; Chao, Z.C.; Potter, S.M. Long-term activity-dependent plasticity of action potential propagation delay and amplitude in cortical networks. PLoS ONE 2008, 3, e2088. [Google Scholar] [CrossRef] [PubMed]
- Lapicque, L. Quantitative investigations of electrical nerve excitation treated as polarization. 1907. Biol. Cybern. 2007, 97, 341–349. [Google Scholar] [PubMed]
- Weiss, G. Sur la possibilite de rendre comparables entre eux les appareils servant a l’excitation electrique. Arch. Ital. Biol. 1901, 35, 413–446. [Google Scholar]
- Cox, D.R. The regression analysis of binary sequences. J. R. Stat. Soc. Ser. B (Methodological) 1958, 20, 215–242. [Google Scholar]
- Tehovnik, E.J.; Tolias, A.S.; Sultan, F.; Slocum, W.M.; Logothetis, N.K. Direct and indirect activation of cortical neurons by electrical microstimulation. J. Neurophysiol. 2006, 96, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Ranck, J.B., Jr. Which elements are excited in electrical stimulation of mammalian central nervous system: A review. Brain Res. 1975, 98, 417–440. [Google Scholar] [CrossRef]
- Histed, M.H.; Bonin, V.; Reid, R.C. Direct activation of sparse, distributed populations of cortical neurons by electrical microstimulation. Neuron 2009, 63, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Rattay, F. The basic mechanism for the electrical stimulation of the nervous system. Neuroscience 1999, 89, 335–346. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuykendal, M.L.; Potter, S.M.; Grover, M.A.; DeWeerth, S.P. Targeted Stimulation Using Differences in Activation Probability across the Strength–Duration Space. Processes 2017, 5, 14. https://doi.org/10.3390/pr5020014
Kuykendal ML, Potter SM, Grover MA, DeWeerth SP. Targeted Stimulation Using Differences in Activation Probability across the Strength–Duration Space. Processes. 2017; 5(2):14. https://doi.org/10.3390/pr5020014
Chicago/Turabian StyleKuykendal, Michelle L., Steve M. Potter, Martha A. Grover, and Stephen P. DeWeerth. 2017. "Targeted Stimulation Using Differences in Activation Probability across the Strength–Duration Space" Processes 5, no. 2: 14. https://doi.org/10.3390/pr5020014





