RNA-Seq as an Emerging Tool for Marine Dinoflagellate Transcriptome Analysis: Process and Challenges
Abstract
:1. Introduction
2. Dinoflagellates Transcriptomics: A Brief History
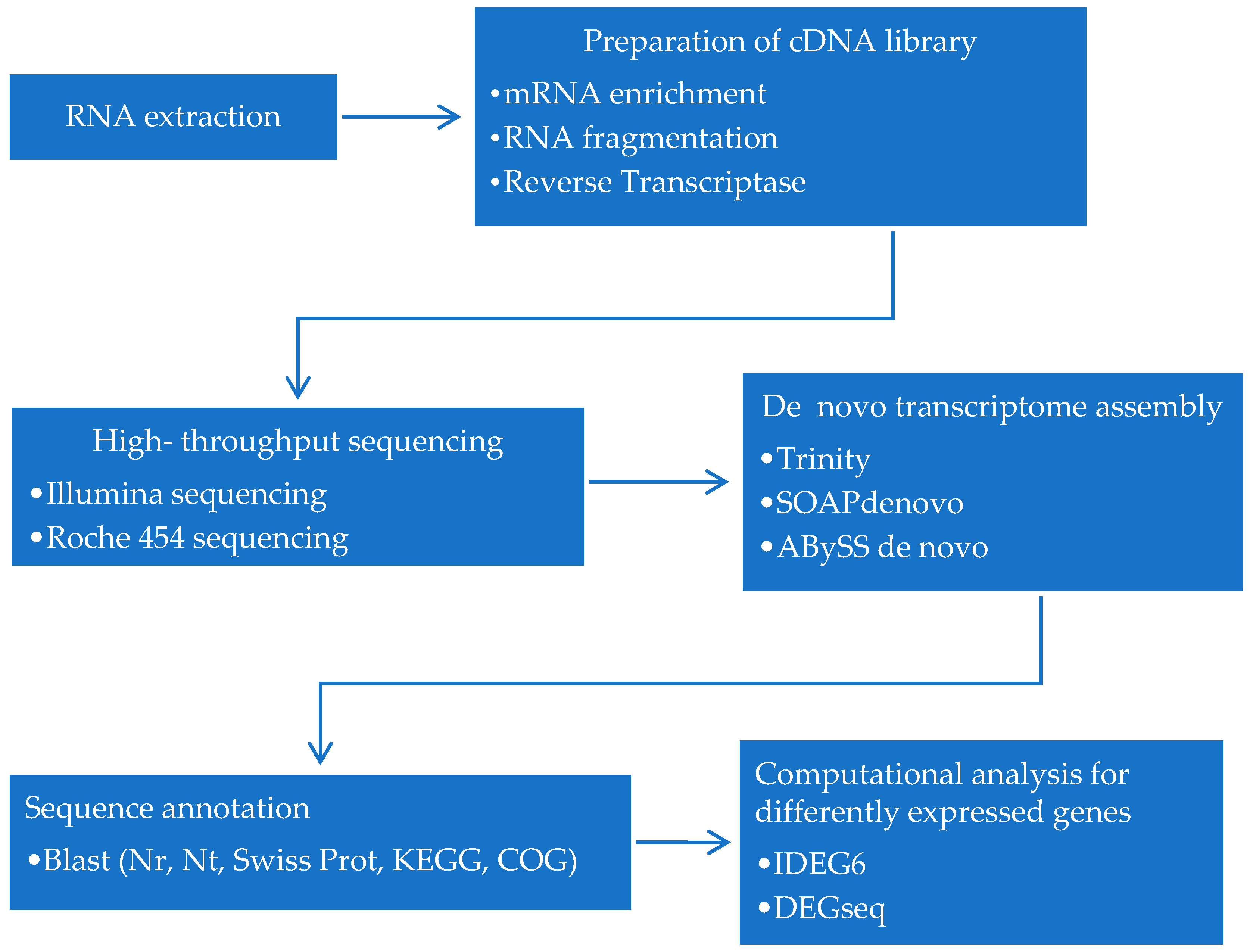
3. Strategies for Dinoflagellates RNA-Seq
3.1. Isolation of RNA
3.2. Preparation of cDNA Library
3.3. High-Throughput Sequencing
3.4. De Novo Transcriptome Assembly
3.5. Sequence Annotation
3.6. Computational Analysis for Differently Expressed Genes
4. Challenges and Consideration in Dinoflagellates Transcriptome Studies Using RNA-Seq
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cooper, J.T.; Sinclair, G.A.; Wawrik, B. Transcriptome analysis of Scrippsiella trochoidea CCMP 3099 reveals physiological changes related to nitrate depletion. Front. Microbiol. 2016, 7, 639. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, A.; Evans, A.N.; Kulis, D.M.; Hackett, J.D.; Erdner, D.L.; Anderson, D.M.; Bhattacharya, D. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS ONE 2010, 5, e9688. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Beszteri, S.; Tillmann, U.; Cembella, A.; John, U. Growth-and nutrient-dependent gene expression in the toxigenic marine dinoflagellate Alexandrium minutum. Harmful Algae 2011, 12, 55–69. [Google Scholar] [CrossRef] [Green Version]
- Baumgarten, S.; Bayer, T.; Aranda, M.; Liew, Y.J.; Carr, A.; Micklem, G.; Voolstra, C.R. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genom. 2013, 14, 704. [Google Scholar] [CrossRef] [PubMed]
- Perini, F.; Casabianca, A.; Battocchi, C.; Accoroni, S.; Totti, C.; Penna, A. New approach using the real-time PCR method for estimation of the toxic marine dinoflagellate Ostreopsis cf. ovata in marine environment. PLoS ONE 2011, 6, e17699. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Malkiel, E.; Katz, J.; Adolf, J.E.; Place, A.R. A dinoflagellate exploits toxins to immobilize prey prior to ingestion. Proc. Natl. Acad. Sci. USA 2010, 107, 2082–2087. [Google Scholar] [CrossRef] [PubMed]
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 16, 551–569. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1. [Google Scholar] [CrossRef] [PubMed]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. Genet. 2009, 10, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Ba, Y.; Zhuang, Q.; Zhong, G. RNA-Seq technology and its application in fish transcriptomics. OMICS J. Integr. Biol. 2014, 18, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Gen. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Bayer, T.; Aranda, M.; Sunagawa, S.; Yum, L.K.; DeSalvo, M.K.; Lindquist, E.; Coffroth, M.A.; Voolstra, C.R.; Medina, M. Symbiodinium transcriptomes: Genome insights into the dinoflagellate symbionts of reef-building corals. PLoS ONE 2012, 7, e35269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, S.-F.; Lin, L.; Wang, D.-Z. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar. Drugs 2014, 12, 5698–5718. [Google Scholar] [CrossRef] [PubMed]
- Gierz, S.L.; Forêt, S.; Leggat, W. Transcriptomic analysis of thermally stressed Symbiodinium reveals differential expression of stress and metabolism genes. Front. Plant Sci. 2017, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.E.; Pepper, A.E.; Campbell, L. De novo assembly and characterization of the transcriptome of the toxic dinoflagellate Karenia brevis. BMC Genom. 2014, 15, 888. [Google Scholar] [CrossRef] [PubMed]
- Pawlowiez, R.; Morey, J.S.; Darius, H.T.; Chinain, M.; Van Dolah, F.M. Transcriptome sequencing reveals single domain Type I-like polyketide synthases in the toxic dinoflagellate Gambierdiscus polynesiensis. Harmful Algae 2014, 36, 29–37. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Whole Transcriptomic Analysis Provides Insights into Molecular Mechanisms for Toxin Biosynthesis in a Toxic Dinoflagellate Alexandrium catenella (ACHK-T). Toxins 2017, 9, 213. [Google Scholar] [CrossRef] [PubMed]
- Rosic, N.; Ling, E.Y.S.; Chan, C.-K.K.; Lee, H.C.; Kaniewska, P.; Edwards, D.; Dove, S.; Hoegh-Guldberg, O. Unfolding the secrets of coral-Algal symbiosis. ISME J. 2015, 9, 844–856. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Lin, X.; Li, L.; Li, M.; Palenik, B.; Lin, S. Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 2017, 11, 2209–2218. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Zhang, H.; Hannick, L.; Lin, S. Metatranscriptome profiling reveals versatile N-nutrient utilization, CO 2 limitation, oxidative stress, and active toxin production in an Alexandrium fundyense bloom. Harmful Algae 2015, 42, 60–70. [Google Scholar] [CrossRef]
- Ogura, A.; Lin, M.; Shigenobu, Y.; Fujiwara, A.; Ikeo, K.; Nagai, S. Effective gene collection from the metatranscriptome of marine microorganisms. BMC Genom. 2011, 12, S15. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Thomson, N.R. Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 2010, 13, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Tjaden, B. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol. 2015, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.A.; Ponnala, L.; Weber, C.A. Strategies for transcriptome analysis in nonmodel plants. Am. J. Bot. 2012, 99, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Taroncher-Oldenburg, G.; Anderson, D.M. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 2000, 66, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Hosoi-Tanabe, S.; Tomishima, S.; Nagai, S.; Sako, Y. Identification of a gene induced in conjugation-promoted cells of toxic marine dinoflagellates Alexandrium tamarense and Alexandrium catenella using differential display analysis. FEMS Microbiol. Lett. 2015, 251, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Karako-Lampert, S.; Hershkovits, G.; Stambler, N.; Simon-Blecher, N.; Achituv, Y.; Dubinsky, Z.; Katcoff, D.J. Differential gene expression in Symbiodinium microadriaticum clade B following stress. Mar. Biotechnol. 2006, 8, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Sturtevant, J. Applications of differential-display reverse transcription-PCR to molecular pathogenesis and medical mycology. Clin. Microbiol. Rev. 2000, 13, 408–427. [Google Scholar] [CrossRef] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S.; Johnson, M.; Bridgham, J.; Golda, G.; Lloyd, D.H.; Johnson, D.; Roth, R. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000, 18, 630. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, O.K.; Hastings, J. Novel dinoflagellate clock-related genes identified through microarray analysis. J. Phycol. 2003, 39, 519–526. [Google Scholar] [CrossRef]
- Van Dolah, F.M.; Lidie, K.B.; Morey, J.S.; Brunelle, S.A.; Ryan, J.C.; Monroe, E.A.; Haynes, B.L. Microarray Analysis of Diurnal-and Circadian-Regulated Genes in the Florida Red-Tide Dinoflagellate Karenia Brevis (Dinophyceae). J. Phycol. 2007, 43, 741–752. [Google Scholar] [CrossRef]
- Yang, I.; John, U.; Beszteri, S.; Glöckner, G.; Krock, B.; Goesmann, A.; Cembella, A.D. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genom. 2010, 11, 248. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Selander, E.; Pavia, H.; John, U. Grazer-induced toxin formation in dinoflagellates: A transcriptomic model study. Eur. J. Phycol. 2011, 46, 66–73. [Google Scholar] [CrossRef]
- Johnson, J.G.; Janech, M.G.; Van Dolah, F.M. Caspase-like activity during aging and cell death in the toxic dinoflagellate Karenia brevis. Harmful Algae 2014, 31, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Coyne, K.J.; Burkholder, J.M.; Feldman, R.A.; Hutchins, D.A.; Cary, S.C. Modified serial analysis of gene expression method for construction of gene expression profiles of microbial eukaryotic species. Appl. Environ. Microbiol. 2004, 70, 5298–5304. [Google Scholar] [CrossRef] [PubMed]
- Erdner, D.L.; Anderson, D.M. Global transcriptional profiling of the toxic dinoflagellate Alexandrium fundyense using massively parallel signature sequencing. BMC Genom. 2016, 7, 88. [Google Scholar] [CrossRef]
- Leggat, W.; Hoegh-Guldberg, O.; Dove, S.; Yellowlees, D. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals. J. Phycol. 2007, 43, 1010–1021. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Stüken, A.; Orr, R.J.S.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morey, J.S.; Monroe, E.A.; Kinney, A.L.; Beal, M.; Johnson, J.G.; Hitchcock, G.L.; Van Dolah, F.M. Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genom. 2011, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Fung-Leung, W.P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS ONE 2014, 9, e78644. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Wakatsuki, T.; Hada, A.; Ryo, A. Use of serial analysis of gene expression (SAGE) technology. J. Immunol. Methods 2001, 250, 45–66. [Google Scholar] [CrossRef]
- Mutz, K.O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Harlow, L.D.; Koutoulis, A.; Hallegraeff, G.M. A novel, simplified technique for preservation and rapid isolation of total RNA from the toxic dinoflagellate Alexandrium catenella (Dinophyceae). Phycologia 2006, 45, 311–318. [Google Scholar] [CrossRef]
- Rosic, N.N.; Hoegh-Guldberg, O. A method for extracting a high-quality RNA from Symbiodinium sp. J. Appl. Phycol. 2010, 22, 139–146. [Google Scholar] [CrossRef]
- Nagaraj, S.H.; Gasser, R.B.; Ranganathan, S. A hitchhiker’s guide to expressed sequence tag (EST) analysis. Brief. Bioinform. 2007, 8, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, J.; Lundeberg, J. The plasticity of the mammalian transcriptome. Genomics 2010, 95, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lowe, C.D.; Mello, L.V.; Samatar, N.; Martin, L.E.; Montagnes, D.J.S.; Watts, P.C. The transcriptome of the novel dinoflagellate Oxyrrhis marina (Alveolata: Dinophyceae): Response to salinity examined by 454 sequencing. BMC Genom. 2011, 12, 519. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Corey, D.R. RNA sequencing: Platform selection, experimental design, and data interpretation. Nucleic Acid Ther. 2012, 22, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Law, M. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012, 2012, 251364. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Misra, R.V.; Dallman, T.J.; Constantinidou, C.; Gharbia, S.E.; Wain, J.; Pallen, M.J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012, 30, 434–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blythe, M.J.; Kao, D.; Malla, S.; Rowsell, J.; Wilson, R.; Evans, D.; Aboobaker, A.A. A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS ONE 2010, 5, e15617. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Gen. 2010, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Gen. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Hamada, M. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Cai, M. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Levin, R.A.; Beltran, V.H.; Hill, R.; Kjelleberg, S.; McDougald, D.; Steinberg, P.D.; Van Oppen, M.J.H. Sex, Scavengers, and Chaperones: Transcriptome Secrets of Divergent Symbiodinium Thermal Tolerances. Mol. Biol. Evol. 2016, 33, 2201–2215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hou, Y.; Miranda, L.; Campbell, D.A.; Sturm, N.R.; Gaasterland, T.; Lin, S. Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. USA 2007, 104, 4618–4623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dungan, C.F.; Lin, S. Introns, alternative splicing, spliced leader trans-splicing and differential expression of pcna and cyclin in Perkinsus marinus. Protist 2011, 162, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sui, Z.; Chang, L.; Kang, K.; Ma, J.; Kong, F.; Zhou, W.; Wang, J.; Guo, L.; Geng, H.; et al. Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the Illumina platform. Gene 2014, 537, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Koid, A.E.; Terrado, R.; Campbell, V.; Caron, D.A.; Heidelberg, K.B. Changes in gene expression of Prymnesium parvum induced by nitrogen and phosphorus limitation. Front. Microbiol. 2015, 6, 631. [Google Scholar] [CrossRef]
- Jang, S.H.; Jeong, H.J.; Chon, J.K.; Lee, S.Y. De novo assembly and characterization of the transcriptome of the newly described dinoflagellate Ansanella granifera: Spotlight on flagellum-associated genes. Mar. Genom. 2016, 33, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Chen, Z. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wohlrab, S.; Groth, M.; Glöckner, G.; Guillou, L.; John, U. Transcriptomic profiling of Alexandrium fundyense during physical interaction with or exposure to chemical signals from the parasite Amoebophrya. Mol. Ecol. 2016, 25, 1294–1307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, R.; Wang, H.; Suh, Y.S.; Ki, J.-S. Transcriptomic profiles reveal the genome-wide responses of the harmful dinoflagellate Cochlodinium polykrikoides when exposed to the algicide copper sulfate. BMC Genom. 2016, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Palumbi, S.R. Lineage-specific transcriptional profiles of Symbiodinium spp. Unaltered by heat stress in a coral host. Mol. Biol. Evol. 2014, 31, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Gibson, A.K.; Smith, Z.; Fuqua, C.; Clay, K.; Colbourne, J.K. Why so many unknown genes? Partitioning orphans from a representative transcriptome of the lone star tick Amblyomma americanum. BMC Genom. 2013, 14, 135. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.M.; Rödelsperger, C.; Eichholz, K.; Tillmann, U.; Cembella, A.; McGaughran, A.; John, U. Transcriptomic characterisation and genomic glimps into the toxigenic dinoflagellate Azadinium spinosum, with emphasis on polykeitde synthase genes. BMC Genom. 2015, 16, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morey, J.S.; Van Dolah, F.M. Global analysis of mRNA half-lives and de novo transcription in a dinoflagellate, Karenia brevis. PLoS ONE 2013, 8, e66347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baudrimont, A.; Voegeli, S.; Viloria, E.C.; Stritt, F.; Lenon, M.; Wada, T.; Becskei, A. Multiplexed gene control reveals rapid mRNA turnover. Sci. Adv. 2017, 3, e1700006. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, A.H. Next generation sequencing of microbial transcriptomes: Challenges and opportunities. FEMS Microbiol. Lett. 2009, 302, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Brunelle, S.A.; Van Dolah, F.M. Post-transcriptional Regulation of S-Phase Genes in the Dinoflagellate, Karenia brevis. J. Eukaryot. Microbiol. 2011, 58, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Qiu, L.; Hou, Z.; Zhang, Q.; Wu, J.; Gao, Q.; Song, L. Computational identification of microRNAs from the expressed sequence tags of toxic dinoflagellate Alexandrium tamarense. Evol. Bioinform. 2013, 9, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Sui, Z.; Zhang, S.; Du, Q.; Ren, Y.; Liu, Y.; Kong, F.; Zhong, J.; Ma, Q. Identification of microRNAs in the toxigenic Dinoflagellate Alexandrium catenella by high-throughput illumina sequencing and bioinformatic analysis. PLoS ONE 2015, 10, e0138709. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Z.; Zhang, H.; Zhang, Y.; Zhang, S.F. Marine dinoflagellate proteomics: Current status and future perspectives. J. Proteom. 2014, 105, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.G. Considerations for the use of transcriptomics in identifying the ‘genes that matter’ for environmental adaptation. J. Exp. Biol. 2015, 218, 1925–1935. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Pronk, T.E.; van der Veen, J.W.; Ezendam, J.; Van Loveren, H.; Pennings, J.L. Effects of pooling RNA from samples treated with different compounds for determining class specific biomarkers and processes in toxicogenomics. Toxicol. In Vitro 2011, 25, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Jaeckisch, N.; Yang, I.; Wohlrab, S.; Glöckner, G.; Kroymann, J.; Vogel, H.; John, U. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS ONE 2011, 6, e28012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kendziorski, C.; Irizarry, R.A.; Chen, K.S.; Haag, J.D.; Gould, M.N. On the utility of pooling biological samples in microarray experiments. Proc. Natl. Acad. Sci. USA 2005, 102, 4252–4257. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Schrey, A.W.; Richards, C.L. Ten years of transcriptomics in wild populations: What have we learned about their ecology and evolution? Mol. Ecol. 2015, 24, 710–725. [Google Scholar] [CrossRef] [PubMed]
| RNA-Seq | Major Application | Platform | References |
|---|---|---|---|
| Transcriptome sequencing | Transcriptome profiling | Roche 454 | [12] |
| Comprehensive analysis of differently gene expression | Illumina | [13,14] | |
| Single Nucleotide Polymorphisms (SNP) discovery | Illumina | [15] | |
| Discovery and characterization of novel genes | Roche 454 | [16] | |
| Isoform discovery | Illumina | [17] | |
| Pathway analysis | Illumina | [18] | |
| Small RNA sequencing | microRNA profiling | Illumina | [4,19] |
| Gene regulation | Illumina | [19] | |
| Meta-transcriptome sequencing | Analysis of community RNA pool and ecological function of gene | Roche 454 | [20] |
| Community taxonomic structure | Roche 454 | [21] |
| Year | Event | References |
|---|---|---|
| 1995 | Development of microarray method | [29] |
| 1995 | Development of SAGE method | [30] |
| 2000 | Development of MPSS method | [31] |
| 2000 | First DDRT-PCR study to discover differently expressed genes | [25] |
| 2003–2012 | Report on dinoflagellates transcriptome analysis using microarray | [32,33,34,35,36] |
| 2004 | Modified SAGE method for dinoflagellates gene expression analysis introduced | [37] |
| 2006–2010 | Report on dinoflagellates transcriptome analysis using MPSS | [2,38] |
| 2007 | Report on dinoflagellates transcriptome analysis using Expression Sequence Tags (EST) | [39] |
| 2008 | Development RNA-seq method | [40] |
| 2011–2017 | Report on dinoflagellates transcriptome analysis using RNA-seq | [19,41] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, M.A.; Ahmad, A.; Usup, G.; Bunawan, H. RNA-Seq as an Emerging Tool for Marine Dinoflagellate Transcriptome Analysis: Process and Challenges. Processes 2018, 6, 5. https://doi.org/10.3390/pr6010005
Akbar MA, Ahmad A, Usup G, Bunawan H. RNA-Seq as an Emerging Tool for Marine Dinoflagellate Transcriptome Analysis: Process and Challenges. Processes. 2018; 6(1):5. https://doi.org/10.3390/pr6010005
Chicago/Turabian StyleAkbar, Muhamad Afiq, Asmat Ahmad, Gires Usup, and Hamidun Bunawan. 2018. "RNA-Seq as an Emerging Tool for Marine Dinoflagellate Transcriptome Analysis: Process and Challenges" Processes 6, no. 1: 5. https://doi.org/10.3390/pr6010005





