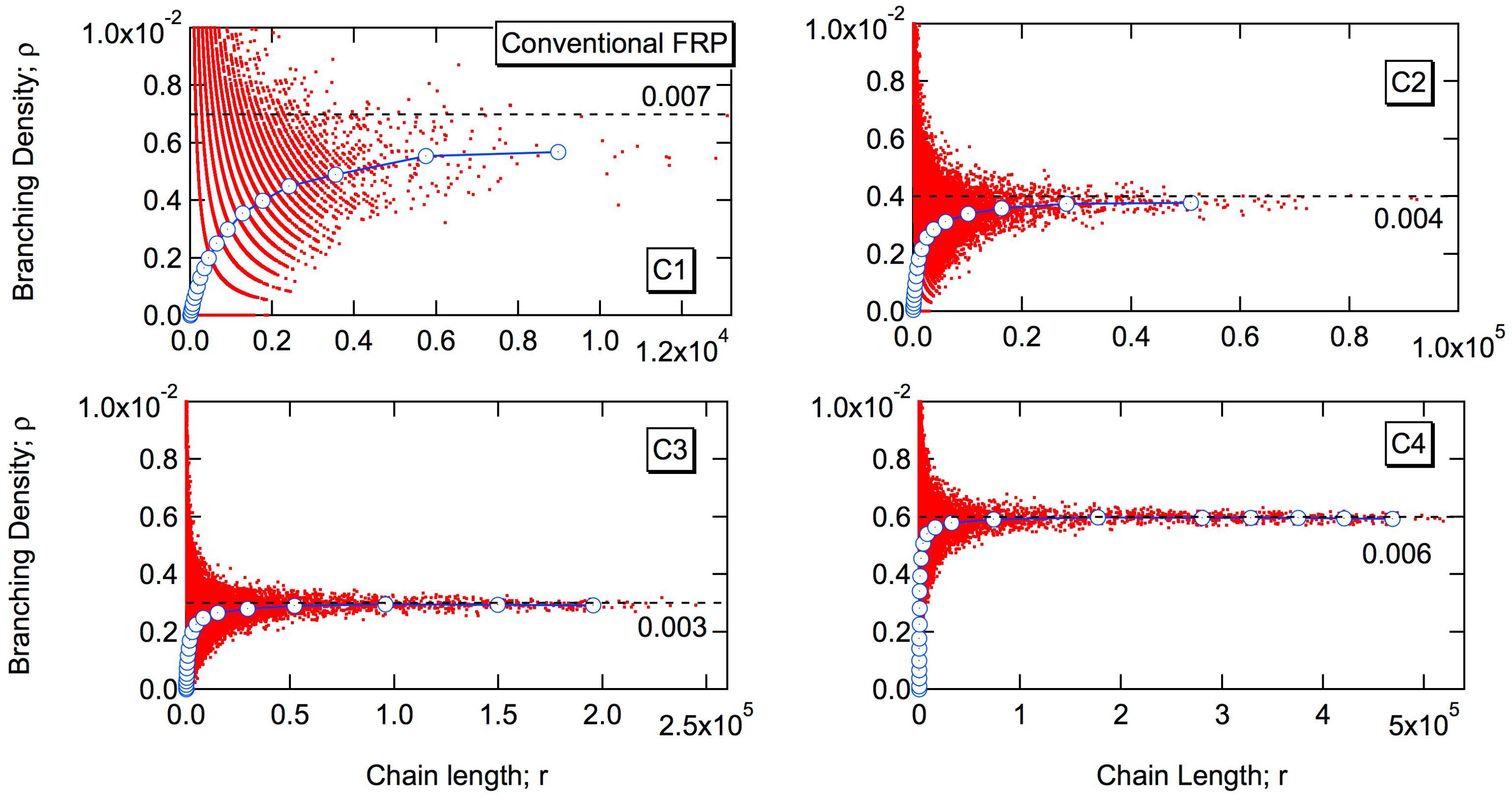
3.1.1. Conventional Free-Radical Emulsion Polymerization
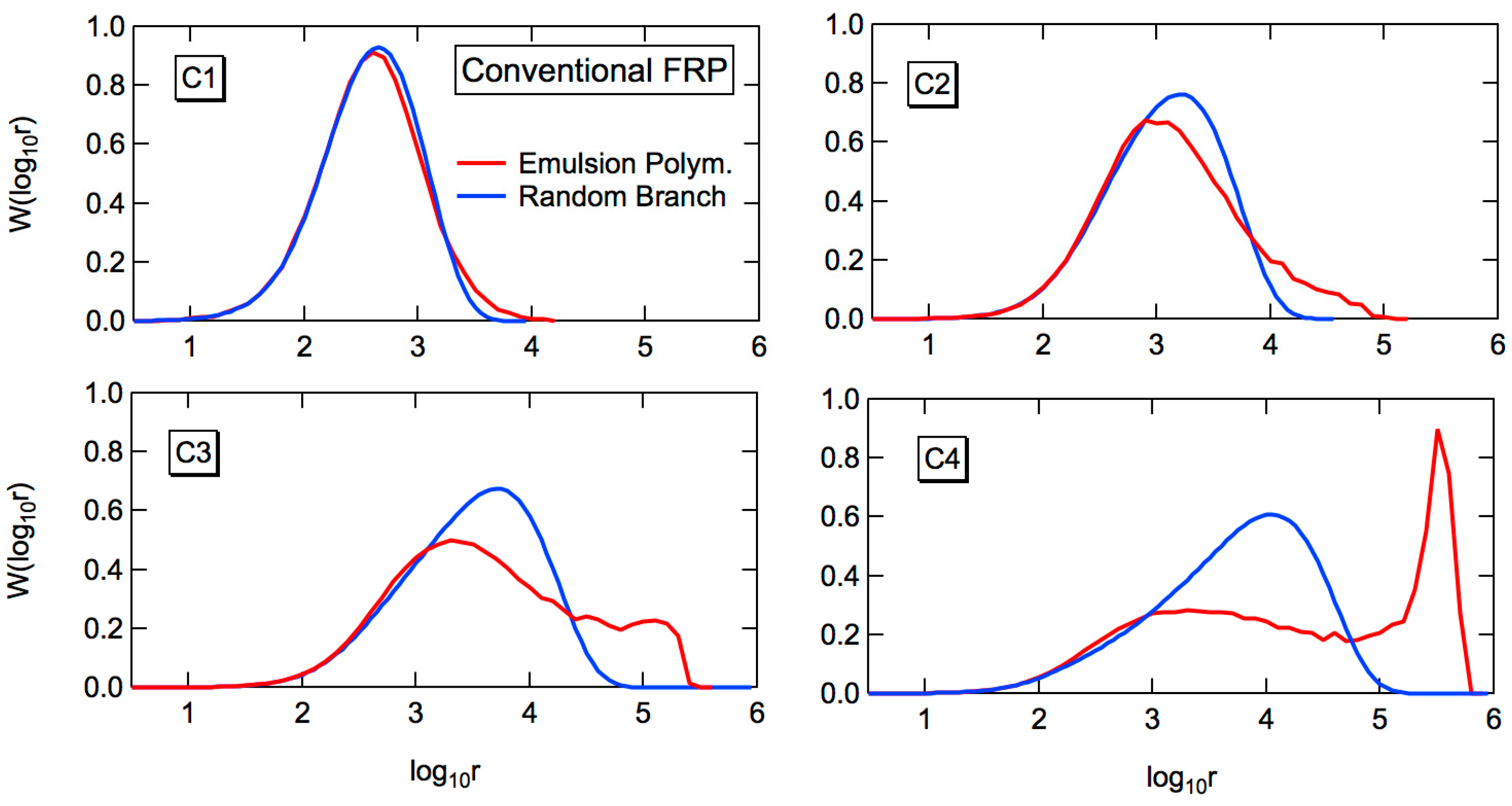
The red curve in
Figure 2 shows the MC simulation results for the weight fraction distribution. The independent variable is the logarithm of chain length, as is usually employed for the GPC measurement. The blue curves in
Figure 2 show the fractional MWDs consisting of
k branch points. The value of
k is 0, 1, 2, 3 from the left to the right. For the second high MW peak shown in C2–C4, the large
k-value fractions are summed up, as shown in the figure. When the branching probability,
Pb is large, a sharp high MW peak appears. In the present condition, the total number of monomeric units incorporated into polymer molecules is,
NM = 1 × 10
6, and therefore, a polymer molecule with log
10r > 6 cannot exist in a polymer particle. The sharp high MW peak is formed due to the limitation of the small particle size. A sharp high MW peak essentially consists of the largest polymer molecule in each polymer particle [
17].
In the present model emulsion polymerization, the weight-average chain length (degree of polymerization),
w is given by [
16,
17]:
and
where
n is the number of primary chains in a particle.
Table 4 shows the comparison of average chain lengths between the MC simulation results and the theoretical values. The errors are rather small, and the accuracy of the present MC simulation is confirmed. Note that the present MC simulation is conducted on the number basis, and therefore, the statistical errors are expected to be smaller for the number-average chain length
n, rather than the weight-average
w.
In order to highlight the characteristics of the emulsion polymerization system, the comparison with the random branched polymer, in which the probability of having a branch point
ρ is the same for monomeric units in polymer, is considered. Suppose we have the primary chains whose number- and weight-average chain lengths are
n,p and
w,p, respectively. When such primary chains are connected through random branching, the number- and weight-average chain length are given, respectively, by [
25]:
Note that Equations (18) and (19) are valid irrespective of the primary chain length distribution.
Comparison of the present model emulsion polymerization and random branching is shown in
Table 5. The number-average chain length
n is the same for both the emulsion polymers and the random branched polymers, because the average branching density is the same. On the other hand, in the case of the emulsion polymers, the expected branching density of the primary chains formed in the earlier stage of polymerization is much larger than those formed in the later stage of polymerization [
5]. The primary chains with larger values of branching density form hubs in the buildup process to connect a large number of primary chains in a polymer molecule, which allows the formation of large sized polymers. It is clearly shown that the weight-average chain length
w is larger for the emulsion polymers, especially for the large branching probability cases.
In the present model emulsion polymerization, the primary chain length distribution follows the most probable distribution, given by Equation (1). When the primary chains conforming to the most probable distribution are connected through random branching, the full weight fraction distribution can be calculated from the following equation [
25]:
where
I1 is the modified Bessel function of the first kind and of the first order.
Figure 3 shows the comparison of the full weight fraction distribution profiles between the emulsion polymers and the random branched polymers. It is clearly shown that the MWD is much broader for the emulsion polymerization.
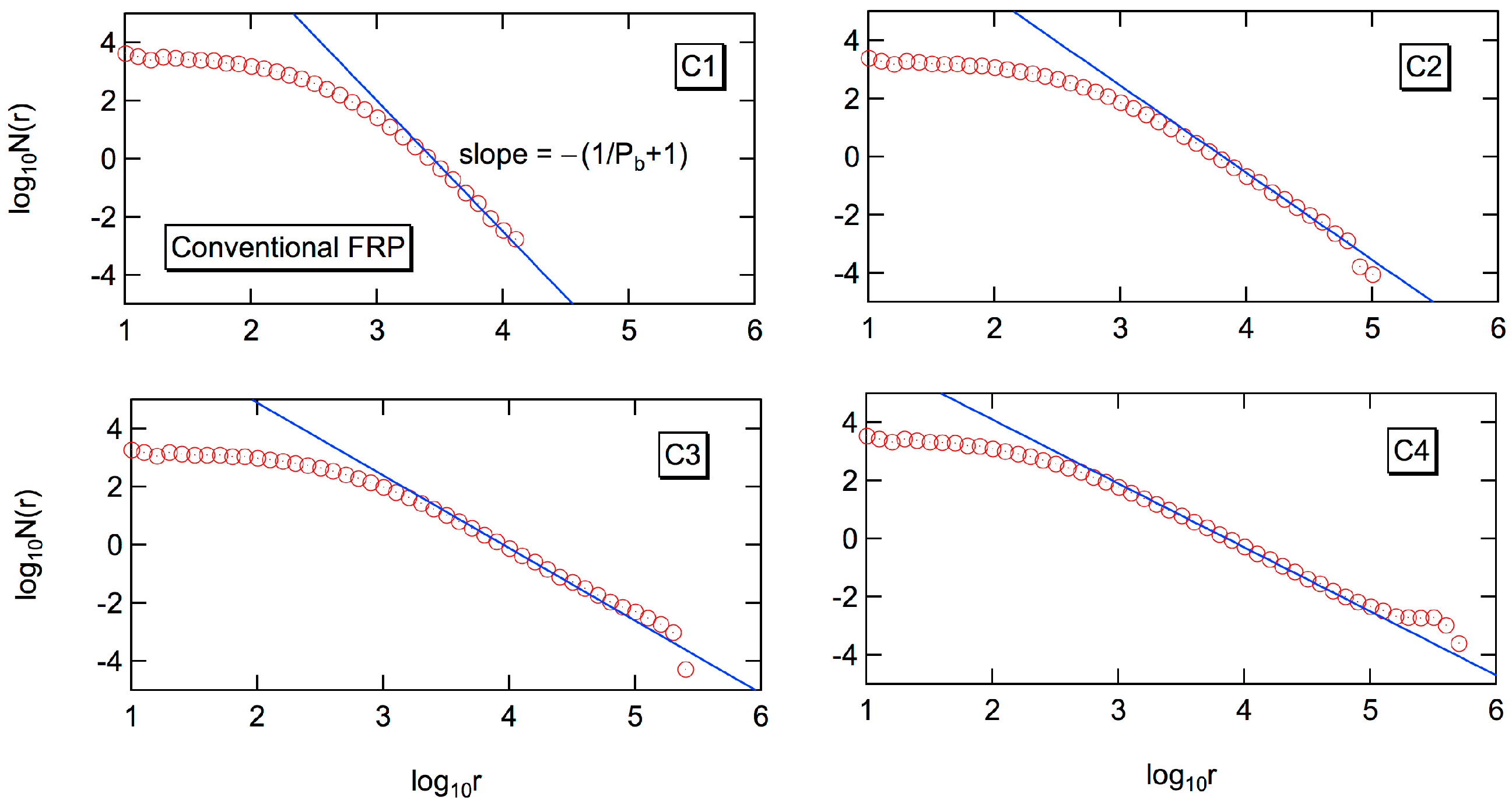
Figure 4 shows the log-log plot of the number fraction distribution,
N(
r). The power law holds for a significant range of chain lengths [
8,
18,
19]. The power exponent for the number fraction distribution,
N(
r) is −(1/
Pb + 1), as shown in the figure. With the relationship,
W(
r) ~
rN(
r), the power exponent of the weight fraction distribution,
W(
r) is −1/
Pb, i.e.,
W(
r) ~
r−α with
α = 1/
Pb. Because the branching probability,
Pb is related with the chain transfer constant
Cfp through Equations (3) and (5), the value of
Cfp could be estimated through the present type of double logarithmic plot, as was illustrated for the emulsion-polymerized polyethylene [
18].
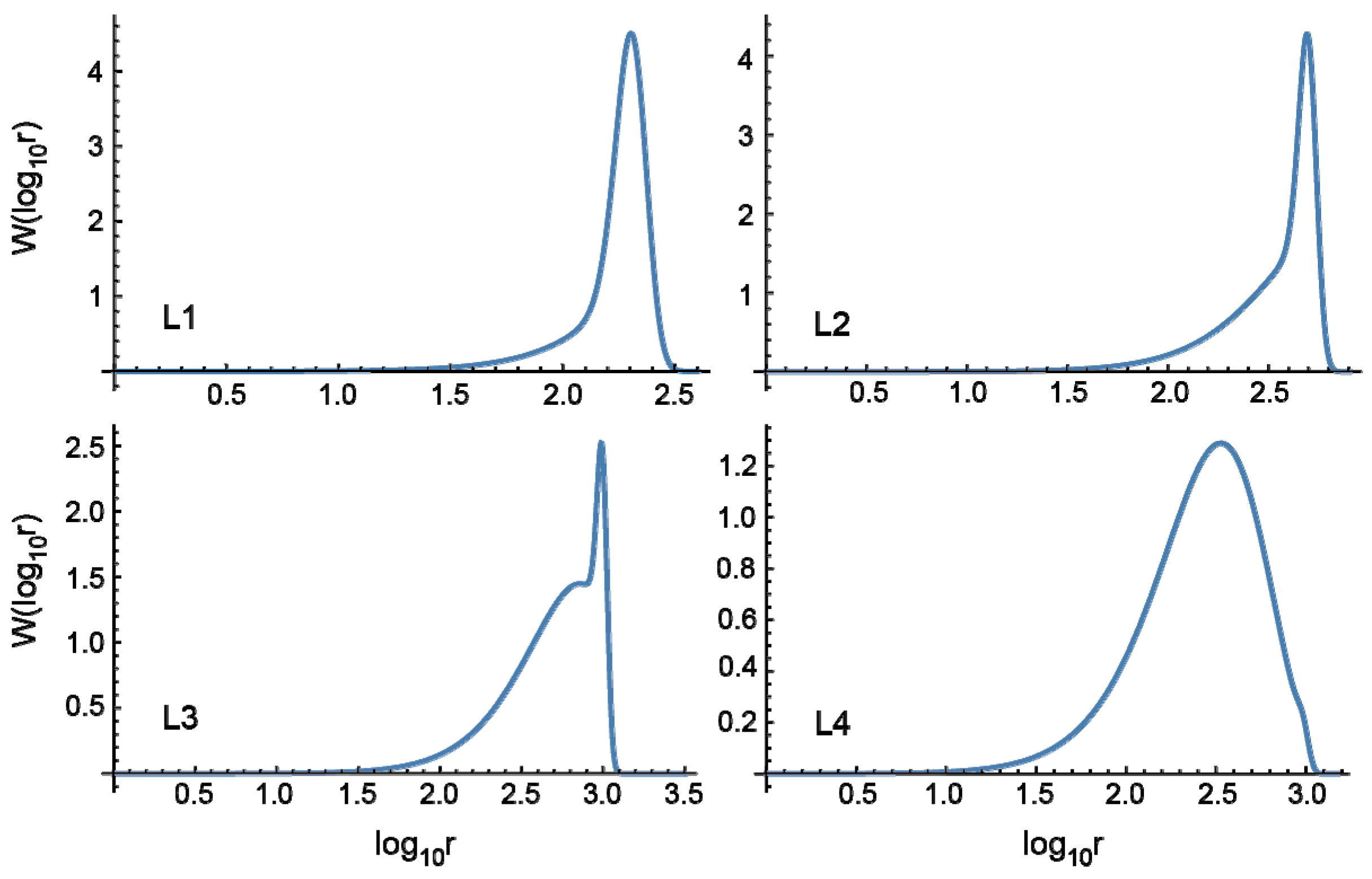
3.1.2. Living Free-Radical Emulsion Polymerization
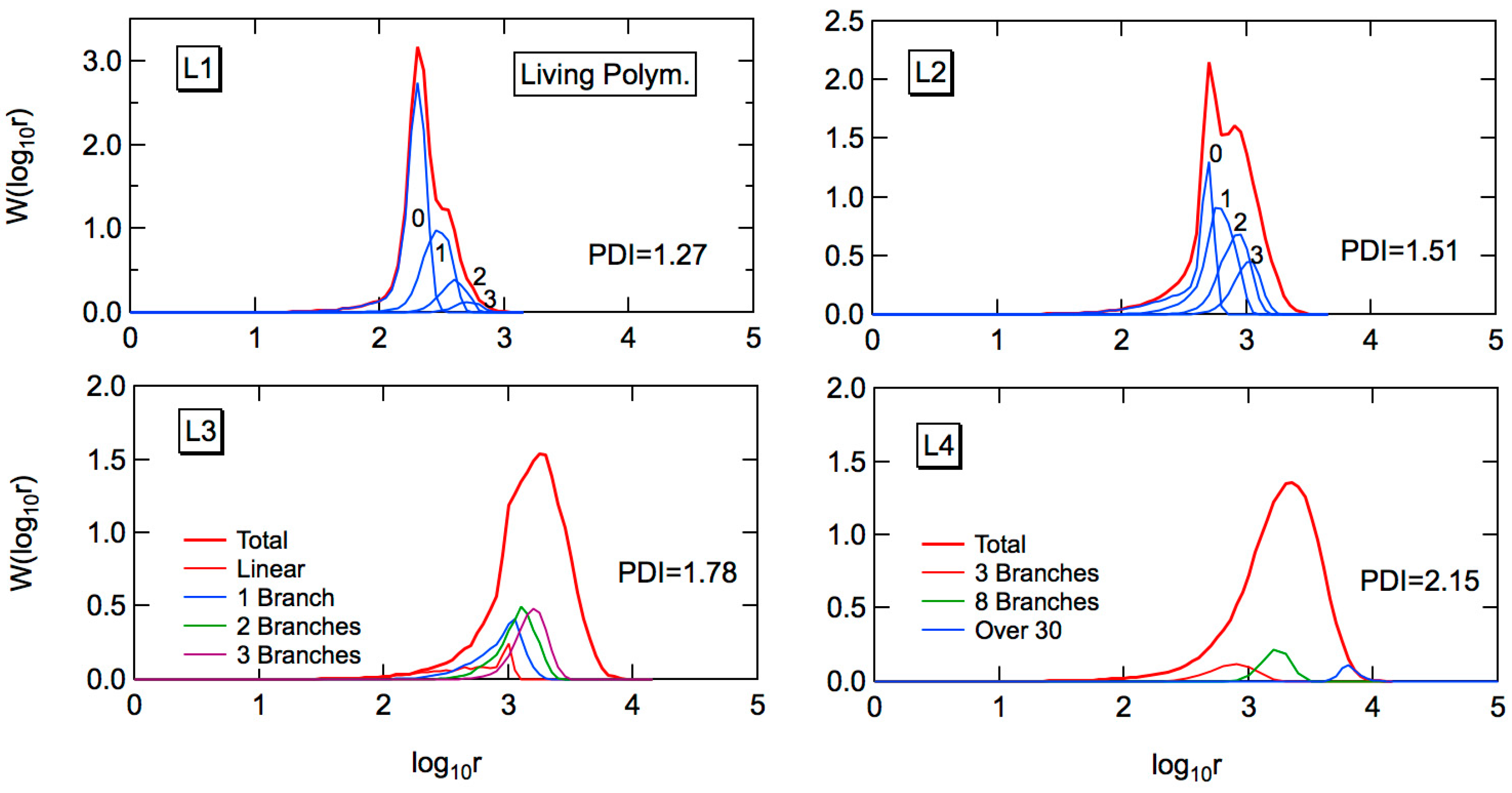
Figure 5 shows the MC simulation results for the full weight fraction distribution, formed through living FRP. A notable difference from the conventional FRP, shown in
Figure 2, is the narrowness of the MWD. Note that the number-average chain length of primary chains
n,p and the average branching density
, as well as the number-average chain lengths of the product polymers
n, are the same for the corresponding polymerization conditions, i.e., C1 and L1, C2 and L2, and so on. Bimodal distributions of
W(log
10r) are shown for L1 and L2. On the other hand, in the present idealized model, a uniform particle size distribution is assumed. In a real system, the MWD is expected to be somewhat broader than the present prediction, and the second peak may be difficult to be observed. In addition, the second peak contains branches and the hydrodynamic volume becomes smaller than the corresponding linear polymers. The peak separation could be difficult in the usual GPC analysis.
The power-law relationship of MWD was found for the conventional emulsion FRP, as was discussed in the previous section. On the other hand, the power law does not hold for the living emulsion polymerization.
In L4, the PDI value is 2.15, while the corresponding conventional FRP gives the PDI value as large as 109.3. In the cases of conventional emulsion FRP, the existence of long-chain branches would be predicted easily judging from a broad MWD. On the other hand, for living emulsion polymerization, the existence of long-chain branches could be overlooked because of a relatively narrow MWD of product polymers.
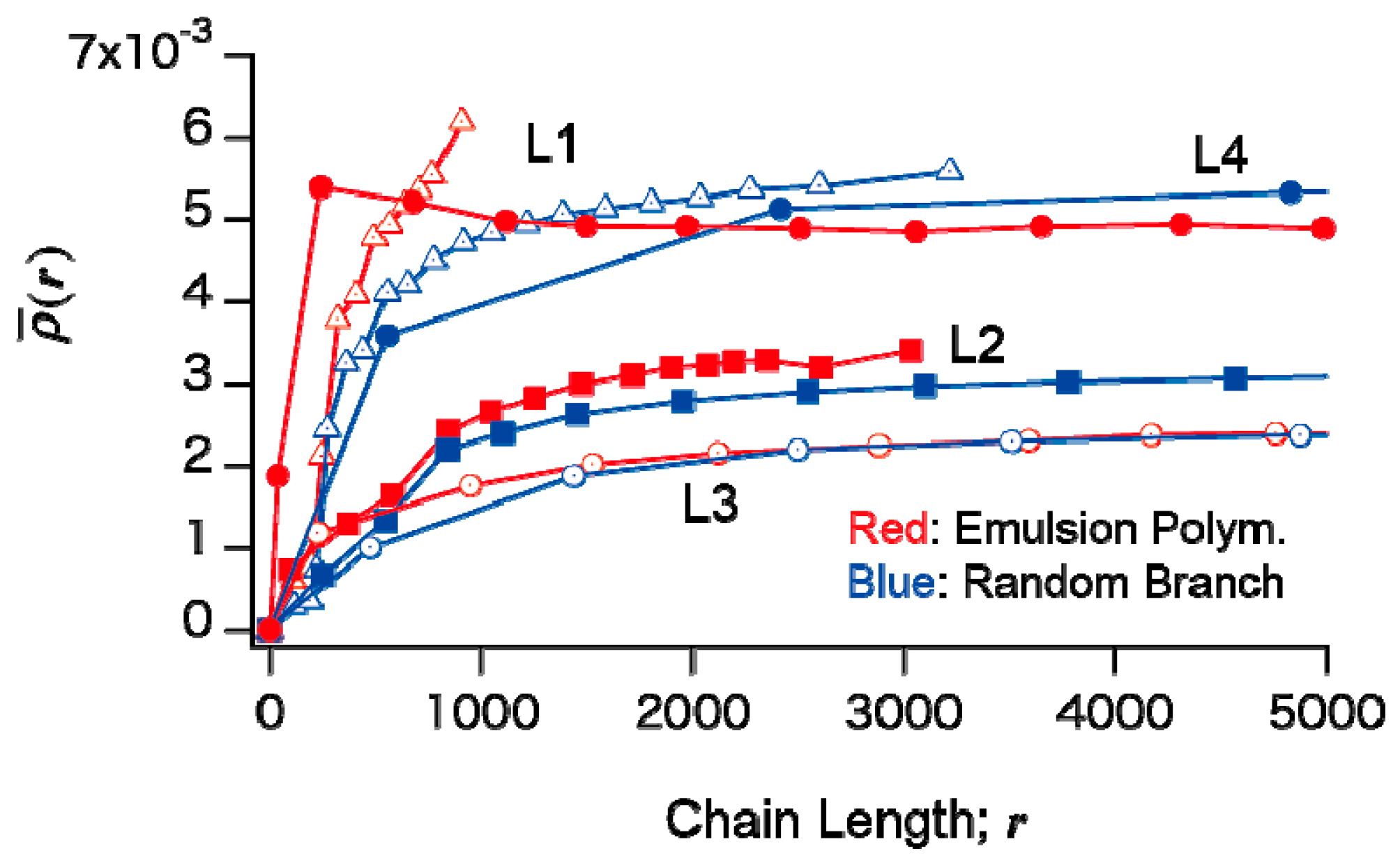
In
Figure 5, the fractional MWDs containing
k branches are also shown. Compared with the fractional MWDs for the conventional FRP shown in
Figure 2, one would notice that the branch points are distributed much more equally for the living emulsion polymerization. Note that the average branching density of the whole system is the same for the corresponding conditions, e.g., C1 and L1, and so on, as shown in
Table 3.
The MWD of linear polymer fraction (k = 0) shows a long tail toward small chain lengths. The long tail is formed through chain transfer to polymer, which causes irreversible deactivation of the chain-end radical. The primary chain length distribution in the present living polymerization system can be calculated theoretically, as follows.
In the case of ideal linear living FRP, where the segment distribution is kept constant throughout the polymerization without termination and chain transfer reactions, the full weight fraction distribution is given by [
23]:
where
F[
a,
b;
x] is the confluent hypergeometric function (Kummer’s function of the first kind), and
z is the average number of active period for a chain that is defined by:
Note that
n,s = 2 in the present set of simulations.
In the present model reaction system, the probability for an active radical to cause chain transfer reaction, resulting in the irreversible termination for the primary chain is equal to
CP, represented by Equation (3), which is kept constant throughout polymerization. When the ideal living chains whose MWD is represented by Equation (21) is subjected to a constant probability of chain transfer, the weight fraction distribution is represented by [
23]:
Equation (23) gives the weight fraction distribution of the primary chains in the present model living emulsion polymerization.
Figure 6 shows the calculated results for L1 to L4. The characteristics of long tails in the low MW region, including a strange distorted fractional MWD profile for
k = 0 of L3 condition in
Figure 5, agree qualitatively with the primary chain length distribution shown in
Figure 6. Note that the primary chains whose distribution is given by
Figure 6 are combined, according to the pertinent emulsion polymerization kinetics, to form the branched polymers whose MWD is shown in
Figure 5.
The number-average chain lengths of the primary chains,
n,p for L1 to L4 are given respectively in
Table 3. The weight-average chain lengths of the primary chains,
w,p can be calculated from Equation (23). On the other hand, if these primary chains are combined to form random branched polymers, the number- and weight-average chain lengths of product polymers can be calculated from Equations (18) and (19). Note that Equations (18) and (19) are valid, irrespective of the primary chain length distribution, as long as the branching is random, i.e., the probability of having a branch point is the same for all units.
Table 6 shows the comparison between the emulsion polymerization and the random branching, in the case of living FRP. The number-average chain length
n is the same for both types of branched polymers, because the average branching density is the same. On the other hand, in the case of living FRP, the branch chains must be formed after the formation of the backbone chain. The branch chain is allowed to grow a shorter period of time than the backbone chain, and is expected to be shorter than the backbone chain. In addition, the branch chains formed later are subjected to branching reaction for a shorter period of time, and therefore, the expected branching density is smaller. On the other hand, in the random branched polymers, the expected branching density is the same for all primary chains and any primary chain can become a branch chain. A long primary chain having large branching density could be connected as a branch chain in the random branching process. Larger polymer molecules can be formed in the random branching process, compared with the living emulsion polymerization, leading to a larger weight-average chain length
w. It is clearly shown that the living emulsion polymerization gives smaller PDI values, indicating narrower MWD.
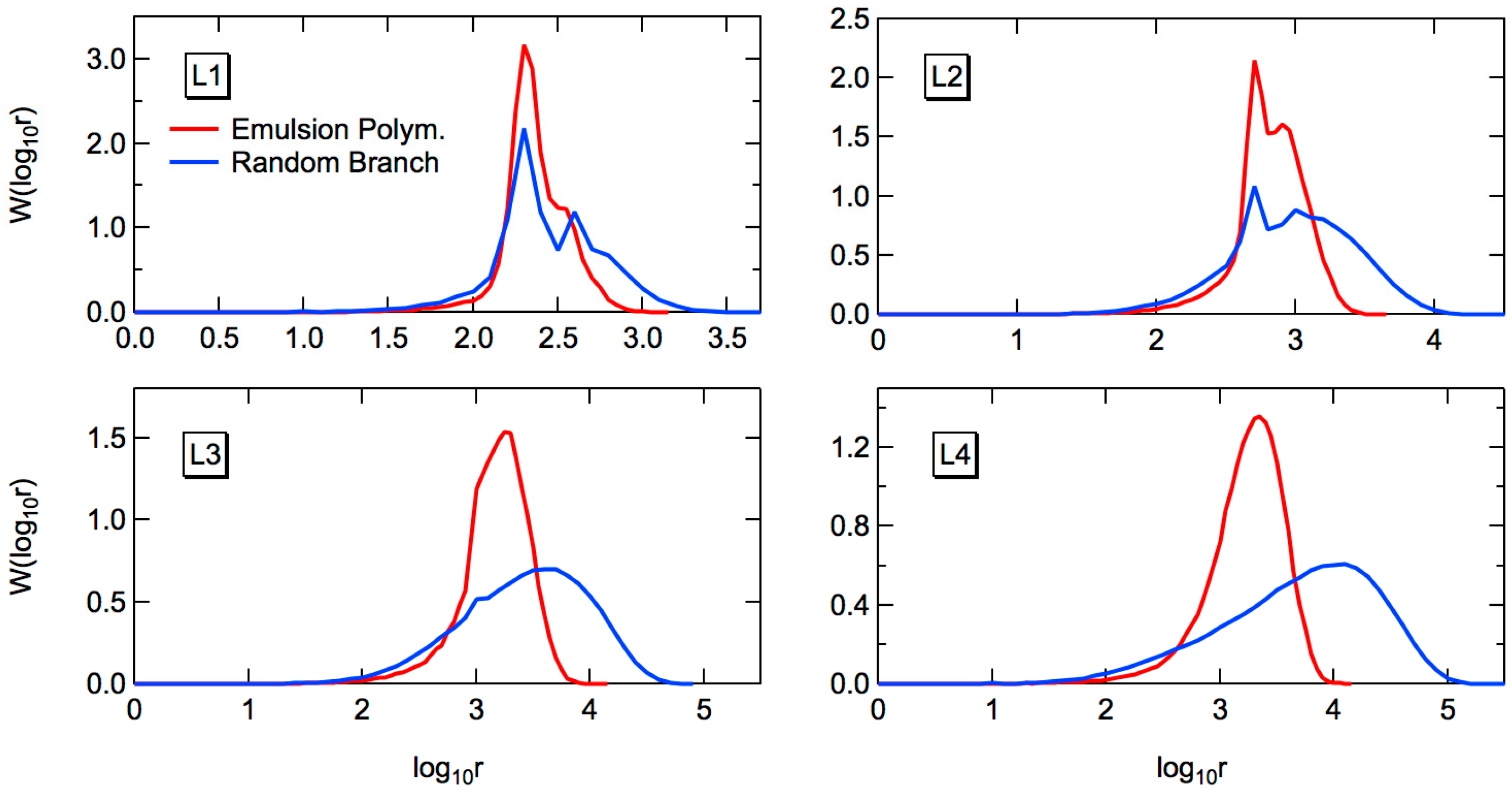
The full weight fraction distribution of the random branched polymers, whose primary chain length distribution is given by Equation (23) can be determined by using the MC simulation.
Figure 7 shows the comparison of weight fraction distribution, between random branching and emulsion polymerization. The formed MWDs in L1–L3 are distorted because of the complicated distribution of the primary chains. Both from the PDI values in
Table 6 and the full MWD profiles in
Figure 7, it can be concluded that the MWD is narrower for the emulsion-polymerized polymers, compared with the corresponding random branched polymers. Remember that for the conventional FRP, the results are totally opposite, i.e., the MWDs are much broader for the emulsion polymers, compared with random branching, in the case of conventional FRP.