Production of Cow’s Milk Free from Beta-Casein A1 and Its Application in the Manufacturing of Specialized Foods for Early Infant Nutrition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
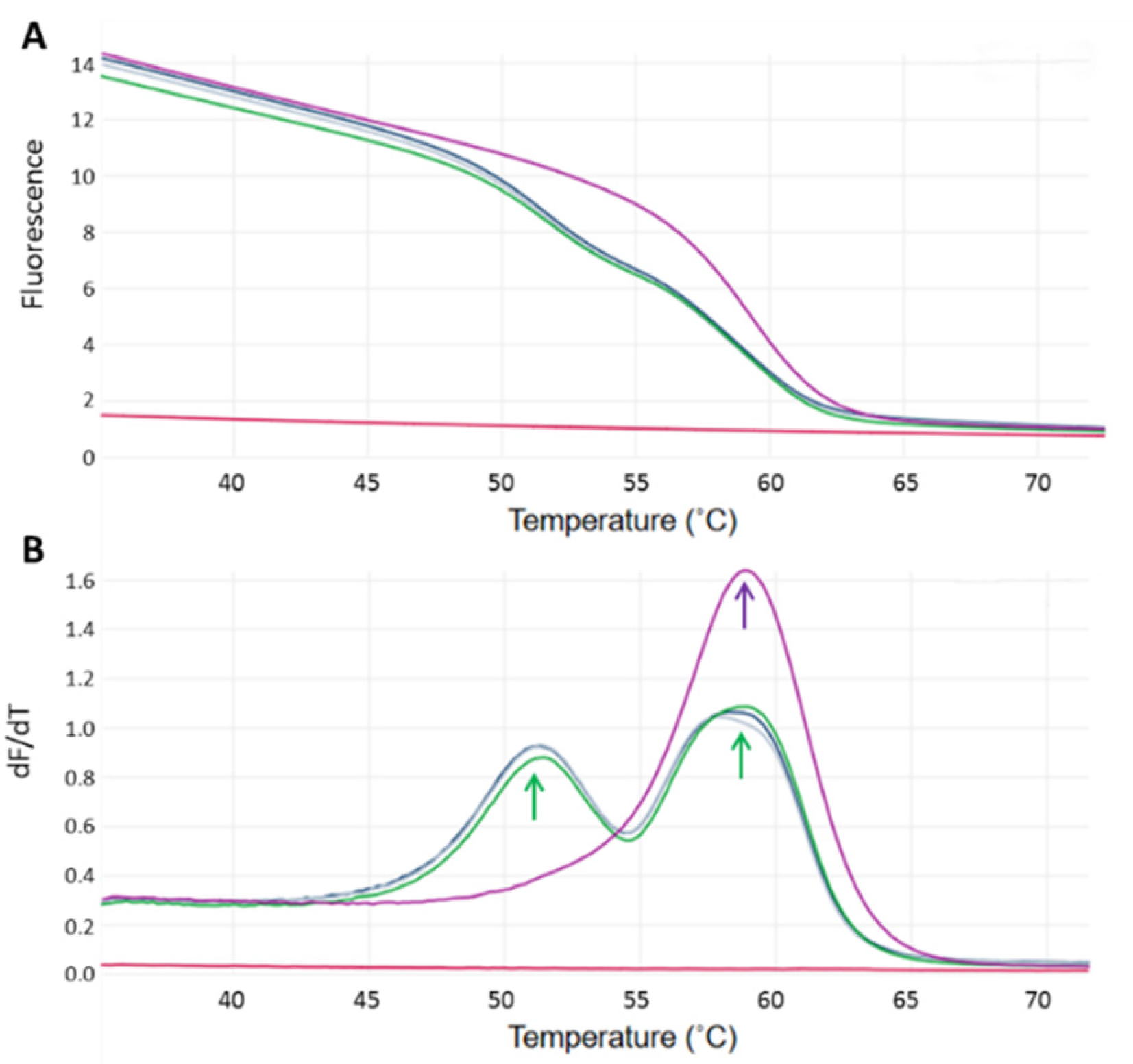
2.2. Genotyping and Selection of Jersey Cows on the Basis of the CSN2 A2 SNP
2.3. Preparation of an IF Based on the BC A2 Milk
2.4. Whole Casein Fractionation from IFs, Human and Raw Cow’s Milk
2.5. BC Polymorphism Phenotyping by HPLC-MS/MS Amino Acid Sequencing
2.6. SGID of Whole Casein Fractions from IFs, Human and Raw Cow’s Milk
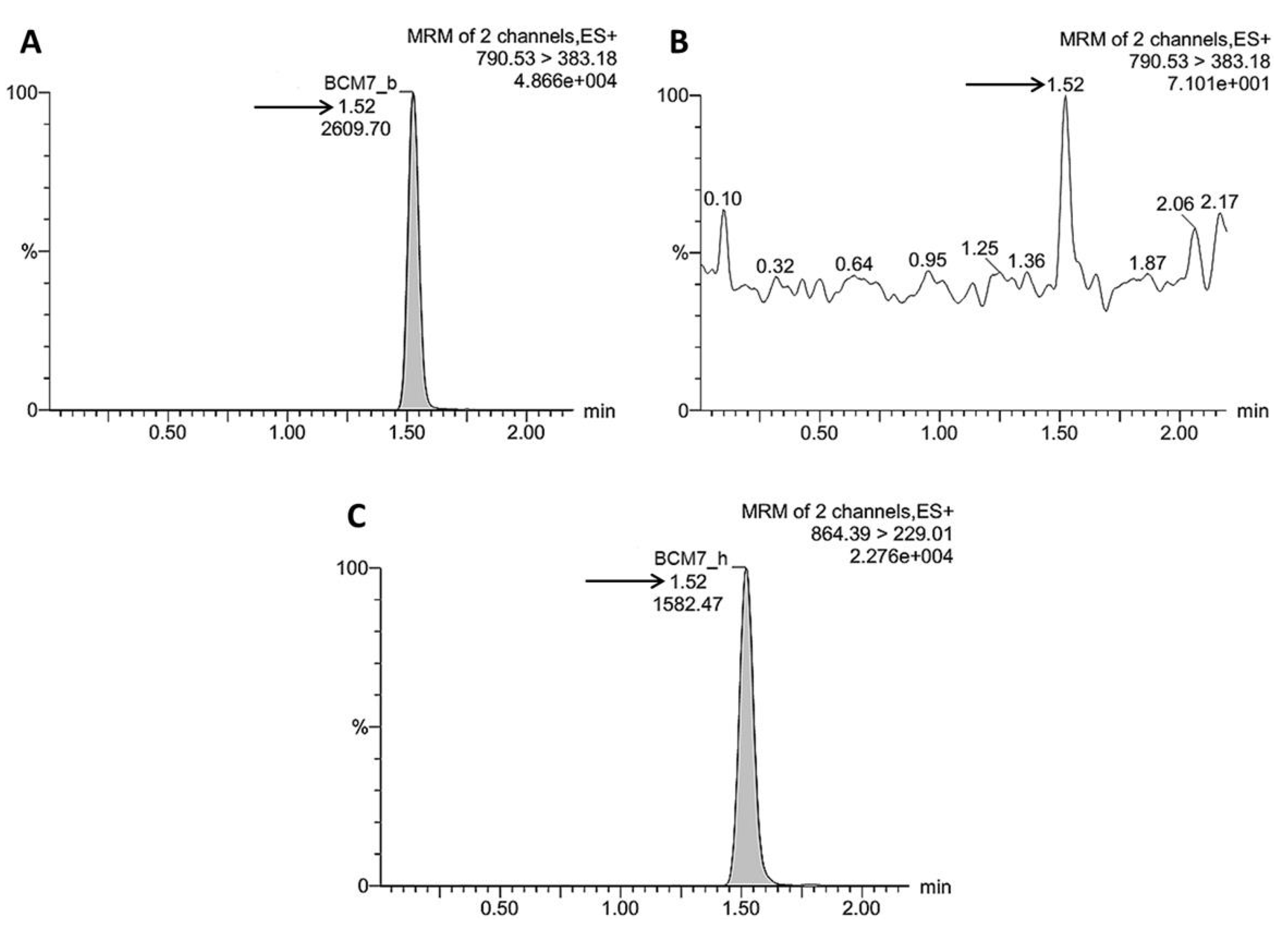
2.7. UPLC–MS/MS Quantification of BCM7 Derived from IFs, Human and Raw Cow’s Milk
2.8. Data Analysis
3. Results
3.1. Genotyping for the CSN2 A2/A1 Alleles in Jersey Cows
3.2. Amino Acid Sequence and Relative Abundance of BC A2 and BC A1 in Whole Casein Fractions from IFs and Raw Cow’s Milk
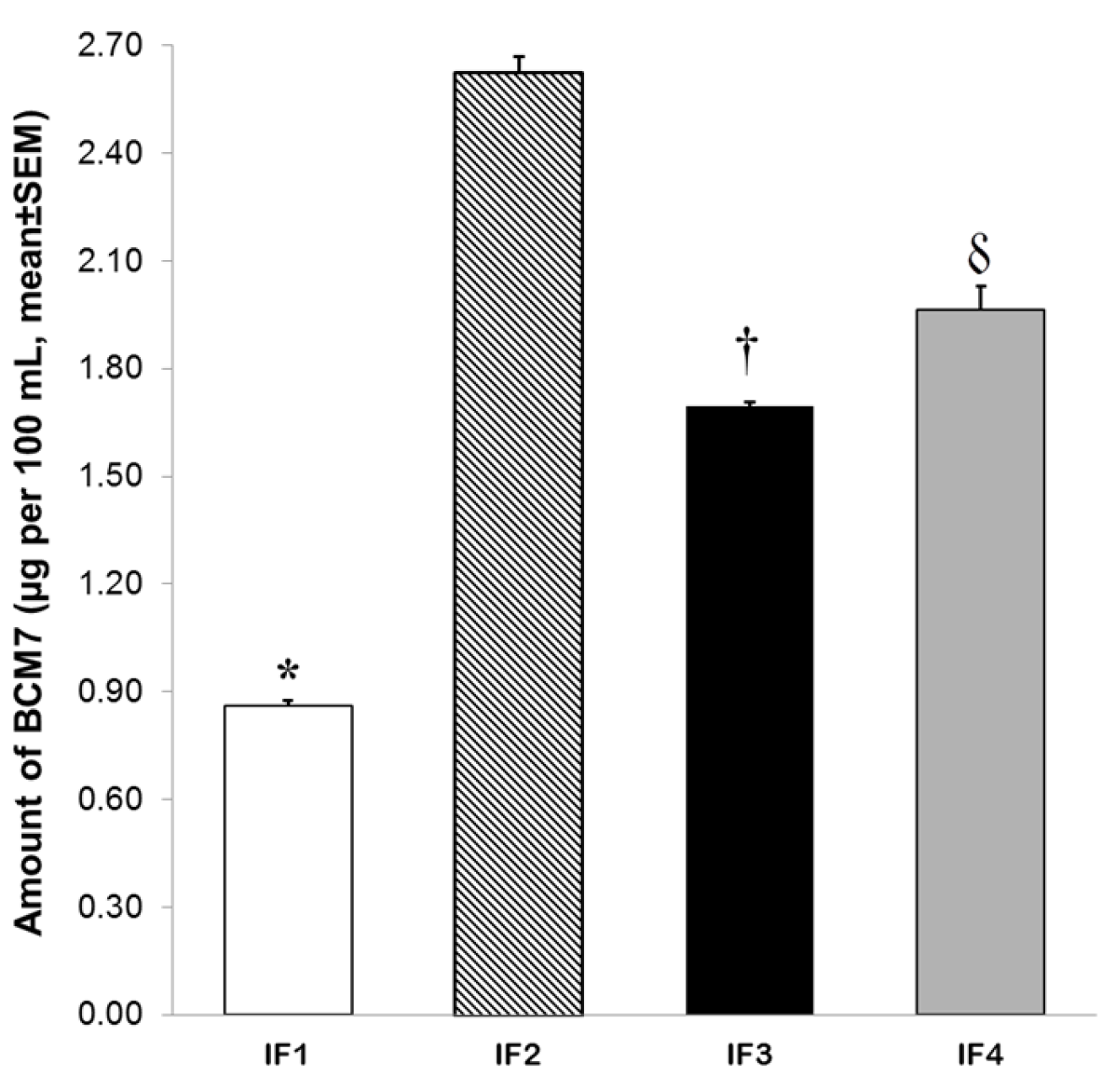
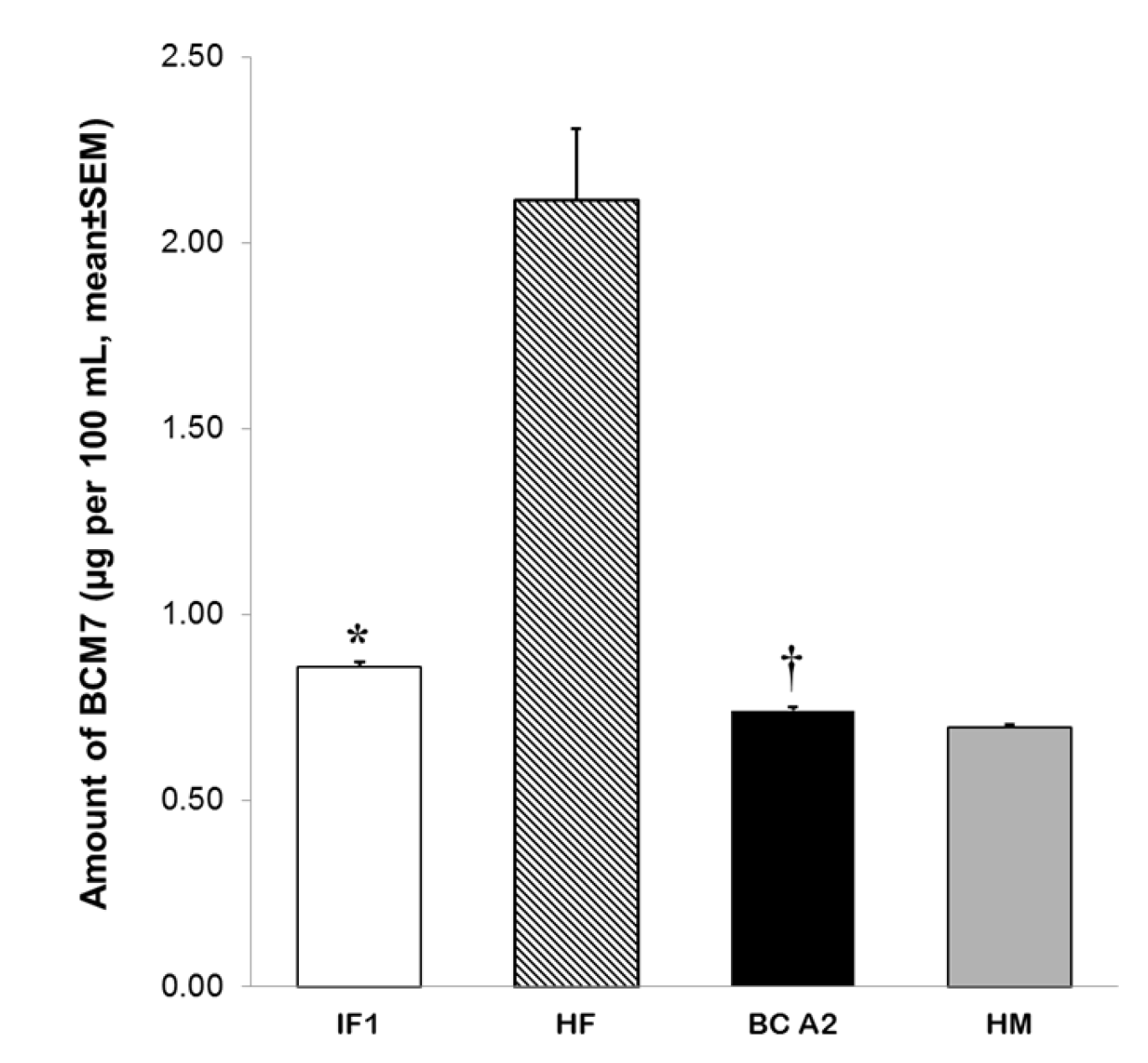
3.3. Release of BCM7 from IFs, Human and Cow’s Milk Following SGID
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Korhonen, H. Milk-derived bioactive peptides: From science to applications. J. Funct. Foods 2009, 1, 177–187. [Google Scholar] [CrossRef]
- Mulvihill, D.M.; Ennis, M.P. Functional milk proteins: Production and utilization. In Advanced Dairy Chemistry Proteins, 3rd ed.; Fox, P.F., McSweeney, P.L.H., Eds.; Springer International Publishing AG: New York, NY, USA, 2003; Volume 1, pp. 1175–1228. [Google Scholar]
- Raies, M.H.; Kapila, R.; Shandilya, U.K.; Kapila, S. Impact of milk derived β-casomorphins on physiological functions and trends in research: A review. Int. J. Food Prop. 2014, 17, 1726–1741. [Google Scholar]
- Farrell, H.M.; Jimenez-Flores, R.; Bleck, G.T.; Brown, E.M.; Butler, J.E.; Creamer, L.K.; Hicks, C.L.; Hollar, C.M.; Ng-Kwai-Hang, K.F.; Swaisgood, H.E. Nomenclature of the proteins of cows’ milk—Sixth revision. J. Dairy Sci. 2004, 87, 1641–1674. [Google Scholar] [CrossRef]
- Swaisgood, H.E. Chemistry of the caseins. In Advanced Dairy Chemistry, 1st ed.; Fox, P.F, Ed.; Elsevier Applied Science: London, UK, 1992; Volume 1, pp. 63–110. [Google Scholar]
- Kaminski, S.; Cieslinska, A.; Kostyra, E. Polymorphism of bovine beta-casein and its potential effect on human health. J. Appl. Genet. 2007, 48, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, H.; Olsen, H.G.; Hayes, B.; Sehested, E.; Svendsen, M.; Nome, T.; Meuwissen, T.; Lien, S. Casein haplotypes and their association with milk production traits in Norwegian Red cattle. Genet. Sel. Evol. 2009, 41, 24. [Google Scholar] [CrossRef] [PubMed]
- Grosclaude, F.; Mahe, M.F.; Mercier, J.C.; Ribadeau-Dumas, B. Caractérisation des variants génétiques des caséines αsl et β bovines. Eur. J. Biochem. 1972, 26, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, A.; Teschemacher, H.; Lehmann, W.; Gauly, M.; Erhardt, G. Influence of genetic polymorphisms in bovine milk on the occurrence of bioactive peptides. In Proceedings IDF “Milk Protein Polymorphism Seminar II”; International Dairy Federation: Brussels, Belgium, 1997; pp. 459–460. [Google Scholar]
- Jinsmaa, Y.; Yoshikawa, M. Enzymatic release of neocasomorphin and beta-casomorphin from bovine beta-casein. Peptides 1999, 20, 957–962. [Google Scholar] [CrossRef]
- Cieslinska, A.; Kostyra, E.B.; Kostyra, H.; Olenski, K.; Fiedorowicz, E.; Kaminski, S.A. Milk from cows of different β-casein genotypes as a source of β-casomorphin-7. Int. J. Food Sci. Nutr. 2012, 63, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Meisel, H. Chemical characterization and opioid activity of an exorphin isolated from in vivo digests of casein. FEBS Lett. 1986, 196, 223–227. [Google Scholar] [CrossRef]
- Stepnik, M.; Kurek, M. The influence of bovine casein-derived exorphins on mast cells in rodents. Rev. Allergol. 2002, 42, 447–453. [Google Scholar] [CrossRef]
- Kurek, M.; Przybilla, B.; Hermann, K.; Ring, J. A naturally occurring opioid peptide from cow’s milk, beta-casomorphine-7, is a direct histamine releaser in man. Int. Arch. Allergy Immunol. 1992, 97, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Shah, N. Effects of milk-derived bioactives: An overview. Br. J. Nutr. 2000, 84, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Andiran, F.; Dayi, S.; Mete, E. Cows’ milk consumption in constipation and anal fistula in infants and young children. J. Paediatr. Child Health 2003, 39, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.; Woodford, K.; Kukuljan, S.; Pal, S. Comparative Effects of A1 versus A2 Beta-Casein on Gastrointestinal Measures: A Blinded Randomised Cross-over Pilot Study. Eur. J. Clin. Nutr. 2014, 68, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Hedner, J.; Hedner, T. β-Casomorphins induce apnea and irregular breathing in adult rats and newborn rabbits. Life Sci. 1987, 41, 2303–2312. [Google Scholar] [CrossRef]
- Wasilewska, J.; Sienkiewicz-Szlapka, E.; Kuzbida, E.; Jarmolowska, B.; Kaczmarski, M.; Kostyra, E. The exogenous opioid peptides and DPPIV serum activity in infants with apnoea expressed as apparent life threatening events (ALTE). Neuropeptides 2011, 45, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, O.; Kost, N.; Andreeva, O.; Korneeva, E.; Meshavkin, V.; Tarakanova, Y.; Dadayan, A.; Zolotarev, Y.; Grachev, S.; Mikheeva, I.; et al. Autistic children display elevated urine levels of bovine casomorphin-7 immunoreactivity. Peptides 2014, 56, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US) Committee on the Evaluation of the Addition of Ingredients New to Infant Formula. Infant Formula Evaluating the Safety of New Ingredients; The National Academic Press: Washington, DC, USA, 2004. [Google Scholar]
- Kost, N.V.; Sokolov, O.Y.; Kurasova, O.B.; Dmitriev, A.D.; Tarakanova, J.N.; Gabaeva, M.V.; Zolotarev, Y.A.; Dadayan, A.K.; Grachev, S.A.; Korneeva, E.V.; et al. Beta-casomorphins-7 in infants on different type of feeding and different levels of psychomotor development. Peptides 2009, 30, 1854–1860. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, P.J.; Jenni, S.; Fortin, M.G. FRET hybridization probes for the rapid detection of disease resistance alleles in plants: Detection of corky root resistance in lettuce. Mol. Breed. 2004, 13, 323–332. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Garfin, D.E. One-dimensional gel electrophoresis. Methods Enzymol. 2009, 463, 497–513. [Google Scholar] [PubMed]
- Hollar, C.M.; Law, A.J.; Dalgleish, D.G.; Medrano, J.F.; Brown, R.J. Separation of beta-casein A1, A2, and B using cation-exchange fast protein liquid chromatography. J. Dairy Sci. 1991, 74, 3308–3313. [Google Scholar] [CrossRef]
- Lönnerdal, B.; Forsum, E. Casein content of human milk. Am. J. Clin. Nutr. 1985, 41, 113–120. [Google Scholar] [PubMed]
- Andrews, A.T. Proteinases in normal bovine milk and their action on caseins. J. Dairy Res. 1983, 50, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Caliskan, F.; Quintero-Hernández, V.; Restano-Cassulini, R.; Batista, C.V.F.; Zamudio, F.Z.; Coronas, F.I.; Possani, L.D. Turkish scorpion Buthacus macrocentrus: General characterization of the venom and description of Bu1, a potent mammalian Na+-channelα-toxin. Toxicon 2012, 59, 408–415. [Google Scholar] [CrossRef] [PubMed]
- De Noni, I. Release of β-casomorphins 5 and 7 during simulated gastrointestinal digestion of bovine β-casein variants and milk-based infant formulas. Food Chem. 2008, 110, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Raies, M.H.; Kapila, R.; Kapila, S. Release of β-casomorphin-7/5 during simulated gastrointestinal digestion of milk β-casein variants from Indian crossbred cattle (Karan Fries). Food Chem. 2015, 168, 70–79. [Google Scholar]
- Yeh, C.F.; Yang, R.C.; Boyle, T. PopGen32 Program; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Vogel, C.; Marcotte, E.M. Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat. Protoc. 2008, 3, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Sadler, M.J.; Smith, N. Beta-casein proteins and infant growth and development. Infant 2013, 9, 173–176. [Google Scholar]
- Ehrmann, S.; Bartenschlager, H.; Geldermann, H. Quantification of gene effects on single milk proteins in selected groups of dairy cows. J. Anim. Breed. Genet. 1997, 114, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.; Zuniga, L.Z.; Flores, A.R.; Dominguez, R.N.; Valverde, R.R. Polymorphism of three milk protein genes in Mexican Jersey cattle. Elect. J. Biotechnol. 2015, 18, 1–4. [Google Scholar] [CrossRef]
- Mishra, B.P.; Mukesh, M.; Prakash, B.; Sodhi, M.; Kapila, R.; Kishore, A.; Bujarbaruah, K.M. Status of milk protein, β-casein variants among Indian milch animals. Indian J. Anim. Sci. 2009, 79, 722–725. [Google Scholar]
- Çardak, A.D. Effects of genetic variants in milk protein on yield and composition of milk from Holstein-Friesian and Simmentaler cows. S. Afr. J. Agric. Sci. 2005, 35, 41–47. [Google Scholar] [CrossRef]
- Hanusová, E.; Huba, J.; Oravcová, M.; Polák, P.; Vrtková, I. Genetic variants of beta-casein in Holstein dairy cattle in Slovakia. Slovak J. Anim. Sci. 2010, 43, 63–66. [Google Scholar]
- Woodford, K. A2 milk, farmer decisions and risk management. In Proceedings of the 16th International Farm Management Association Congress, Cork University College, Cork, Ireland, 27 July 2007; O’Reilly, S., Keane, M., Enright., P., Eds.; Peer Reviewed Papers Cork University College: Cork, Ireland, 2007; pp. 641–648. [Google Scholar]
- Koletzko, B.; Baker, S.; Cleghorn, G.; Neto, U.F.; Gopalan, S.; Hernell, O.; Hock, Q.S.; Jirapinyo, P.; Lonnerdal, B.; Pencharz, P.; et al. Global standard for the composition of infant formula: Recommendations of an ESPGHAN coordinated international expert group. J. Pediatr. Gastroenterol. Nutr. 2005, 41, 584–599. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Vázquez, M.A. Análisis comparativo de las fórmulas infantiles comercializadas en México. In Fórmulas Infantiles, Nutrición y Tecnología, 1st ed.; Rosado, J.L., Ed.; MGS Ediciones: Querétaro, México, 2017; pp. 303–304. [Google Scholar]
- De Jong, N.; Visser, S.; Olieman, C. Determination of milk proteins by capillary electrophoresis. J. Chromatogr. A 1993, 652, 207–213. [Google Scholar] [CrossRef]
- Heck, J.M.L.; Olieman, C.; Schennink, A.; Van Valenberg, H.J.F.; Visker, M.H.P.W.; Meuldijk, R.C.R.; Van Hooijdonk, A.C.M. Estimation of variation in concentration, phosphorylation and genetic polymorphism of milk proteins using capillary zone electrophoresis. Int. Dairy J. 2008, 18, 548–555. [Google Scholar] [CrossRef]
- Recio, I.; Pérez-Rodríguez, M.L.; Ramos, M.; Amigo, L. Capillary electrophoretic analysis of genetic variants of milk proteins from different species. J. Chromatogr. A 1997, 768, 47–56. [Google Scholar] [CrossRef]
- De Noni, I.; Cattaneo, S. Occurrence of β-casomorphins 5 and 7 in commercial dairy products and in their digests following in vitro simulated gastro-intestinal digestion. Food Chem. 2010, 119, 560–566. [Google Scholar] [CrossRef]
- Cattaneo, S.; Stuknytė, M.; Masotti, F.; De Noni, I. Protein breakdown and release of β-casomorphins during in vitro gastro-intestinal digestion of sterilised model systems of liquid infant formula. Food Chem. 2017, 217, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Cieslinska, A.; Kaminski, S.; Kostyra, E.; Sienkiewicz-Szlapka, E. Betacasomorphin 7 in raw and hydrolyzed milk derived from cows of alternative beta-casein genotypes. Milchwissenschaft 2007, 62, 125–127. [Google Scholar]
- Hernandez-Ledesma, B.; Amigo, L.; Ramos, M.; Recio, I. Release of angiotensin converting enzyme-inhibitory peptides by simulated gastrointestinal digestion of infant formulas. Int. Dairy J. 2004, 14, 889–898. [Google Scholar] [CrossRef]
- Nasripour, A.; Scher, J.; Desorby, S. Baby foods: Formulations and interactions (a review). Crit. Rev. Food Sci. Nutr. 2006, 46, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Ángeles, L.J.L. Formulación de fórmulas infantiles. In Fórmulas Infantiles, Nutrición y Tecnología, 1st ed.; Rosado, J.L., Ed.; MGS Ediciones: Querétaro, México, 2017; pp. 303–304. [Google Scholar]
- Henle, T. Protein-bound advanced glycation end products (AGEs) as bioactive amino acid derivatives in food. Amino Acids 2005, 29, 313–322. [Google Scholar] [CrossRef] [PubMed]
| Sample | Ingredient Composition According to Label | Main Label’s Claim | Protein Content (g/100 mL) | Casein: Whey Protein Ratio (w/w) |
|---|---|---|---|---|
| IF 1 | Beta-casein (BC) A2 skimmed milk, whey protein concentrate, beta-palmitic acid, lutein, choline, DHA, arachidonic acid (ARA), alpha-lactalbumin, prebiotics and nucleotides | With BC A2® milk | 1.33 | 35:65 |
| IF 2 | Skimmed milk, whey protein concentrate, probiotic Lb. reuteri, DHA and ARA | With optimized protein blend Optipro® and Lb. reuteri (LR) | 1.20 | 40:60 |
| IF 3 | Skimmed milk, whey protein concentrate, lutein, DHA, ARA, alpha-lactalbumin and nucleotides | With Biofactors System® | 1.33 | 35:65 |
| IF 4 | Skimmed milk, whey protein concentrate, DHA, ARA, choline, prebiotics and milk fat globule membrane (MFGM) | With DHA, ARA, choline and MFGM for suitable neurodevelopment | 1.35 | 40:60 |
| n | Allele Frequencies | Genotype Frequencies | ||||
|---|---|---|---|---|---|---|
| A2 | A1 | A2A2 | A2A1 | A1A1 | ||
| Cows | 130 | 0.858 | 0.142 | 0.715 | 0.285 | 0 |
| F1 calves | 40 | 1 | 0 | 1 | 0 | 0 |
| F2 calves | 32 | 1 | 0 | 1 | 0 | 0 |
| F3 calves | 25 | 1 | 0 | 1 | 0 | 0 |
| BC Genetic Variant | Sample | |||||
|---|---|---|---|---|---|---|
| IF 1 | IF 2 | IF 3 | IF 4 | Holstein Friesian | BC A2 Milk | |
| BC A2 | 0.864 ± 0.013 | 0.403 ± 0.174 | 0.559 ± 0.011 | 0.497 ± 0.009 | 0.426 ± 0.015 | 0.998 ± 0.008 |
| BC A1 | 0.136 ± 0.010 | 0.597 ± 0.020 | 0.441 ± 0.014 | 0.503 ± 0.011 | 0.574 ± 0.012 | ND |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte-Vázquez, M.Á.; García-Ugalde, C.; Villegas-Gutiérrez, L.M.; García-Almendárez, B.E.; Rosado, J.L. Production of Cow’s Milk Free from Beta-Casein A1 and Its Application in the Manufacturing of Specialized Foods for Early Infant Nutrition. Foods 2017, 6, 50. https://doi.org/10.3390/foods6070050
Duarte-Vázquez MÁ, García-Ugalde C, Villegas-Gutiérrez LM, García-Almendárez BE, Rosado JL. Production of Cow’s Milk Free from Beta-Casein A1 and Its Application in the Manufacturing of Specialized Foods for Early Infant Nutrition. Foods. 2017; 6(7):50. https://doi.org/10.3390/foods6070050
Chicago/Turabian StyleDuarte-Vázquez, Miguel Á., Carlos García-Ugalde, Laura M. Villegas-Gutiérrez, Blanca E. García-Almendárez, and Jorge L. Rosado. 2017. "Production of Cow’s Milk Free from Beta-Casein A1 and Its Application in the Manufacturing of Specialized Foods for Early Infant Nutrition" Foods 6, no. 7: 50. https://doi.org/10.3390/foods6070050





