Transcriptional Suppression of Renal Antioxidant Enzyme Systems in Guinea Pigs Exposed to Polymerized Cell-Free Hemoglobin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Antibodies
2.2. Surgical Preparation and Experimental Protocol
2.3. Design of PCR Primers and TaqMan Probes for Quantitative Gene Expression Assay
2.4. RNA Extraction
2.5. Real-Time Quantitative-PCR (qPCR)
2.6. Antioxidant Enzyme Activity Assays
2.7. Immunohistochemistry
2.8. CpG Island Prediction and Primer Design for Promoter Methylation PCR
2.9. Analysis of DNA Methylation
2.10. Lipid Peroxidation Assay
2.11. Statistical Analysis
3. Results
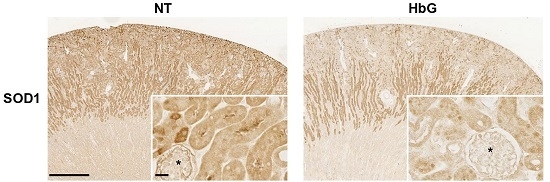
3.1. Reduced Renal SOD, GPx, and CAT Transcription, Expression, and Activity Following HbG Infusion
3.2. DNA Promoter Analysis: HbG-Transfusion Induces DNA Hypermethylation of SOD, GPx, and CAT
3.3. Increased Renal Lipid Peroxidation Following HbG Transfusion
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HBOC: | Hemoglobin-based oxygen carrier |
| HbG: | Oxyglobin/polymerized bovine hemoglobin |
| ET: | Exchange transfusion |
| ROS: | Reactive oxygen species |
| SOD: | Superoxide dismutase |
| GPx: | Glutathione peroxidase |
| CAT: | Catalase |
| NT: | No treatment |
| 4-HNE: | 4-hydroxynonenal |
| bp: | Base pairs |
References
- Alayash, A.I. Blood substitutes: Why Haven’t We been More Successful? Trends Biotechnol. 2014, 32, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Buehler, P.W.; D’Agnillo, F.; Schaer, D.J. Hemoglobin-based oxygen carriers: From Mechanisms of Toxicity and Clearance to Rational Drug Design. Trends Mol. Med. 2010, 16, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Silverman, T.A.; Weiskopf, R.B. Hemoglobin-based oxygen carriers: Current Status and Future Directions. Anesthesiology 2009, 111, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Butt, O.I.; Buehler, P.W.; D’Agnillo, F. Blood-brain barrier disruption and oxidative stress in guinea pig after systemic exposure to modified cell-free hemoglobin. Am. J. Pathol. 2011, 178, 1316–1328. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.H.; Zhang, X.; Williams, M.C.; Hicks, W.; Buehler, P.W.; D’Agnillo, F. Sodium nitrite potentiates renal oxidative stress and injury in hemoglobin exposed guinea pigs. Toxicology 2015, 333, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Pisoschi, A.M.; Pop, A. The role of antioxidants in the chemistry of oxidative stress: A Review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Rosen, D.G.; Zhou, Y.; Feng, L.; Yang, G.; Liu, J.; Huang, P. Mitochondrial manganese-superoxide dismutase expression in ovarian cancer: Role in Cell Proliferation and Response to Oxidative Stress. J. Biol. Chem. 2005, 280, 39485–39492. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.R.; Hitchler, M.J.; Domann, F.E. Regulation of SOD2 in cancer by histone modifications and CpG methylation: Closing the Loop between Redox Biology and Epigenetics. Antioxid. Redox Signal. 2013, 18, 1946–1955. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Naushad, S.M.; Reddy, C.A.; Rupasree, Y.; Pavani, A.; Digumarti, R.R.; Gottumukkala, S.R.; Kuppusamy, P.; Kutala, V.K. Cross-talk between one-carbon metabolism and xenobiotic metabolism: Implications on Oxidative DNA Damage and Susceptibility to Breast Cancer. Cell. Biochem. Biophys. 2011, 61, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Divyya, S.; Naushad, S.M.; Murthy, P.V.; Reddy Ch, R.; Kutala, V.K. GCPII modulates oxidative stress and prostate cancer susceptibility through changes in methylation of RASSF1, BNIP3, GSTP1 and Ec-SOD. Mol. Biol. Rep. 2013, 40, 5541–5550. [Google Scholar] [CrossRef] [PubMed]
- Teoh-Fitzgerald, M.L.; Fitzgerald, M.P.; Jensen, T.J.; Futscher, B.W.; Domann, F.E. Genetic and epigenetic inactivation of extracellular superoxide dismutase promotes an invasive phenotype in human lung cancer by disrupting ECM homeostasis. Mol. Cancer Res. 2012, 10, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Kulak, M.V.; Cyr, A.R.; Woodfield, G.W.; Bogachek, M.; Spanheimer, P.M.; Li, T.; Price, D.H.; Domann, F.E.; Weigel, R.J. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene 2013, 32, 4043–4051. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.F.; Razvi, M.; Chen, H.; Washington, K.; Roessner, A.; Schneider-Stock, R.; El-Rifai, W. DNA hypermethylation regulates the expression of members of the mu-class glutathione s-transferases and glutathione peroxidases in barrett’s adenocarcinoma. Gut 2009, 58, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Rao, X.; House, M.G.; Nephew, K.P.; Cullen, K.J.; Guo, Z. Gpx3 promoter hypermethylation is a frequent event in human cancer and is associated with tumorigenesis and chemotherapy response. Cancer Lett. 2011, 309, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Min, J.Y.; Lim, S.O.; Jung, G. Downregulation of catalase by reactive oxygen species via hypermethylation of CpG island II on the catalase promoter. FEBS Lett. 2010, 584, 2427–2432. [Google Scholar] [CrossRef] [PubMed]
- Buehler, P.W.; D’Agnillo, F.; Hoffman, V.; Alayash, A.I. Effects of endogenous ascorbate on oxidation, oxygenation, and toxicokinetics of cell-free modified hemoglobin after exchange transfusion in rat and guinea pig. J. Pharmacol. Exp. Ther. 2007, 323, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, I.B. Evolution and the biosynthesis of ascorbic acid. Science 1973, 182, 1271–1272. [Google Scholar] [CrossRef] [PubMed]
- Butt, O.I.; Buehler, P.W.; D’Agnillo, F. Differential induction of renal heme oxygenase and ferritin in ascorbate and nonascorbate producing species transfused with modified cell-free hemoglobin. Antioxid. Redox Signal. 2010, 12, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Buehler, P.W.; Boykins, R.A.; Jia, Y.; Norris, S.; Freedberg, D.I.; Alayash, A.I. Structural and functional characterization of glutaraldehyde-polymerized bovine hemoglobin and its isolated fractions. Anal. Chem. 2005, 77, 3466–3478. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Dahiya, R. Methprimer: Designing Primers for Methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [PubMed]
- Takai, D.; Jones, P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA 2002, 99, 3740–3745. [Google Scholar] [CrossRef] [PubMed]
- Vincze, T.; Posfai, J.; Roberts, R.J. Nebcutter: A Program to Cleave DNA with Restriction Enzymes. Nucleic Acids Res. 2003, 31, 3688–3691. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-blast: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cyr, A.R.; Domann, F.E. The redox basis of epigenetic modifications: From Mechanisms to Functional Consequences. Antioxid. Redox Signal. 2011, 15, 551–589. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.J.; Alam, J.; Nath, K.A. Physiology and pathophysiology of heme: Implications for Kidney Disease. J. Am. Soc. Nephrol. 2007, 18, 414–420. [Google Scholar] [CrossRef] [PubMed]
- D’Agnillo, F.; Chang, T.M. Polyhemoglobin-superoxide dismutase-catalase as a blood substitute with antioxidant properties. Nat. Biotechnol. 1998, 16, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Reeder, B.J.; Grey, M.; Silaghi-Dumitrescu, R.L.; Svistunenko, D.A.; Bulow, L.; Cooper, C.E.; Wilson, M.T. Tyrosine residues as redox cofactors in human hemoglobin: Implications for Engineering Nontoxic Blood Substitutes. J. Biol. Chem. 2008, 283, 30780–30787. [Google Scholar] [CrossRef] [PubMed]
- Simoni, J.; Simoni, G.; Moeller, J.F.; Feola, M.; Wesson, D.E. Artificial oxygen carrier with pharmacologic actions of adenosine-5′-triphosphate, adenosine, and reduced glutathione formulated to treat an array of medical conditions. Artif. Organs 2014, 38, 684–690. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L.; Bharani, M.; Pallavi, V. Role of antioxidants and free radicals in health and disease. Adv. Pharmacol. Toxicol. 2006, 7, 29–38. [Google Scholar]
- Aragno, M.; Cutrin, J.C.; Mastrocola, R.; Perrelli, M.G.; Restivo, F.; Poli, G.; Danni, O.; Boccuzzi, G. Oxidative stress and kidney dysfunction due to ischemia/reperfusion in rat: Attenuation by Dehydroepiandrosterone. Kidney Int. 2003, 64, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, Y.; Saari, J.T.; Kang, Y.J. Catalase-overexpressing transgenic mouse heart is resistant to ischemia-reperfusion injury. Am. J. Physiol. 1997, 273, H1090–H1095. [Google Scholar] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Chao, P.C.; Huang, C.N.; Hsu, C.C.; Yin, M.C.; Guo, Y.R. Association of dietary ages with circulating ages, glycated LDL, IL-1alpha and MCP-1 levels in type 2 diabetic patients. Eur. J. Nutr. 2010, 49, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef] [PubMed]
- Abdilla, N.; Tormo, M.C.; Fabia, M.J.; Chaves, F.J.; Saez, G.; Redon, J. Impact of the components of metabolic syndrome on oxidative stress and enzymatic antioxidant activity in essential hypertension. J. Hum. Hypertens. 2007, 21, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.K.; Park, J.S.; Koo, D.B.; Choi, Y.H.; Kim, S.U.; Lee, K.K.; Han, Y.M. Limited demethylation leaves mosaic-type methylation states in cloned bovine pre-implantation embryos. EMBO J. 2002, 21, 1092–1100. [Google Scholar] [CrossRef] [PubMed]
- Gardiner-Garden, M.; Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987, 196, 261–282. [Google Scholar] [CrossRef]
- Xie, X.L.; Yu, Y.; Yuan, Z.F.; Yang, J.; Ma, P.P.; Li, D.C.; Yu, S.K.; An, F.; Feng, X.J.; Zhang, Y. Comparative analysis on content and distribution of CpG sites in milk production traits and mastitis-related genes in dairy cattle. Yi Chuan 2012, 34, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Zhang, R.; Kim, G.Y.; Bae, S.C.; Hyun, J.W. Epigenetic changes induced by oxidative stress in colorectal cancer cells: Methylation of Tumor Suppressor RUNX3. Tumour Biol. 2012, 33, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.C.; Molognoni, F.; Melo, F.H.; Galdieri, L.C.; Carneiro, C.R.; D’Almeida, V.; Correa, M.; Jasiulionis, M.G. Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia 2007, 9, 1111–1121. [Google Scholar] [CrossRef] [PubMed]
- Soberanes, S.; Gonzalez, A.; Urich, D.; Chiarella, S.E.; Radigan, K.A.; Osornio-Vargas, A.; Joseph, J.; Kalyanaraman, B.; Ridge, K.M.; Chandel, N.S.; et al. Particulate matter air pollution induces hypermethylation of the p16 promoter via a mitochondrial ROS-JNK-DNMT1 pathway. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef] [PubMed]





| Gene | Accession Number | Forward Primer Sequence | Reverse Primer Sequence | Probes |
|---|---|---|---|---|
| SOD1 | XM_003467248.3 | GAGACCTGGGCAATGTGACT | GAGAGTCCTCGATGGATACATTGG | CACGCCGTCTGCACCAG |
| SOD2 | XM_003466367.3 | GACAAACCTGAGCCCTAATGGT | AGTCACGTTTGATGGCTTCCA | TTCCCCTTTGGGTTCTC |
| SOD3 | XM_003467399.3 | GCCGCGTCTGGAAACAC | CCTCGCCGGCATGGA | CCGCACTCGATCCTG |
| GPx1 | XM_003476448.3 | CTTCCCGTGCAACCAGTTC | CTTGAGCGAATGCAGAATCTCTTC | ACACCAGGAGAACGCC |
| GPx3 | XM_003464498.3 | GCGAGGAGTACATTCCCTTCAATAA | GCTGGCCACGTTGACAAAA | ACAGAGGCAAATACATCC |
| GPx4 | NM_001256319.1 | CCTTCCCCTGCAACCAGTT | GAACTCCTTGATCTCCTCATTGGT | CCTGGCTCCTGCTTCC |
| CAT | NM_001172925.1 | Applied Biosystems | Cp03755233_m1 | - |
| GAPDH | NM_001172951.1 | Applied Biosystems | Cp03755743_g1 | - |
| Genome | Accession Number | Forward Primer Sequence | Reverse Primer Sequence | Tm (°C) | Size (bp) |
|---|---|---|---|---|---|
| SOD1 | NT_176367.1 | CATCCATCTTGATGGGTCCT | GCGCCGGACTCGATTTA | 57 | 222 |
| SOD2 | NT_176359.1 | GGATCCCTGGGGTGATGTT | GCCCCATAGTCGTAGGGTAA | 58 | 172 |
| SOD3 | NT_176369.1 | GGATGAGGTGGATCTGTCG | GGCATACATGTCTCCGATCT | 57 | 227 |
| GPx1 | NT_176411.1 | CGAGAAGTTGGGGAGGAGTA | CGGACGTACTTGAGCGAATG | 57 | 241 |
| GPx3 | NT_176338.1 | TTCCACCAAAAGTGGGGGTC | GGCCTCAAGCAAGTCTTGGA | 60 | 183 |
| GPx4 | NT_176099.1 | ACCCCTGGCTTGATCGGAAC | CAGCTTCCAGGCACGGTCA | 61 | 153 |
| CAT | NT_176327.1 | GCAGAGTTTGAAGTCGCCTA | ACCTAAAACGCCACACAAAC | 57 | 310 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rentsendorj, O.; Zhang, X.; Williams, M.C.; Buehler, P.W.; D’Agnillo, F. Transcriptional Suppression of Renal Antioxidant Enzyme Systems in Guinea Pigs Exposed to Polymerized Cell-Free Hemoglobin. Toxics 2016, 4, 6. https://doi.org/10.3390/toxics4010006
Rentsendorj O, Zhang X, Williams MC, Buehler PW, D’Agnillo F. Transcriptional Suppression of Renal Antioxidant Enzyme Systems in Guinea Pigs Exposed to Polymerized Cell-Free Hemoglobin. Toxics. 2016; 4(1):6. https://doi.org/10.3390/toxics4010006
Chicago/Turabian StyleRentsendorj, Otgonchimeg, Xiaoyuan Zhang, Matthew C. Williams, Paul W. Buehler, and Felice D’Agnillo. 2016. "Transcriptional Suppression of Renal Antioxidant Enzyme Systems in Guinea Pigs Exposed to Polymerized Cell-Free Hemoglobin" Toxics 4, no. 1: 6. https://doi.org/10.3390/toxics4010006