Insights on Osmotic Tolerance Mechanisms in Escherichia coli Gained from an rpoC Mutation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media and Growth Conditions
2.2. Marker-Less Reconstruction of rpoC Mutation in BW25113
2.3. Growth Kinetic Analysis
2.4. Effects of Amino Acid Supplementation
2.5. Metabolite Analysis
2.6. Effects of Acetic Acid Supplementation
2.7. Cell Membrane Damage Analysis
2.8. Transcriptional Analysis
2.9. Overexpression and Deletion Assay
2.10. Microarray Data Accession Number
3. Results and Discussion
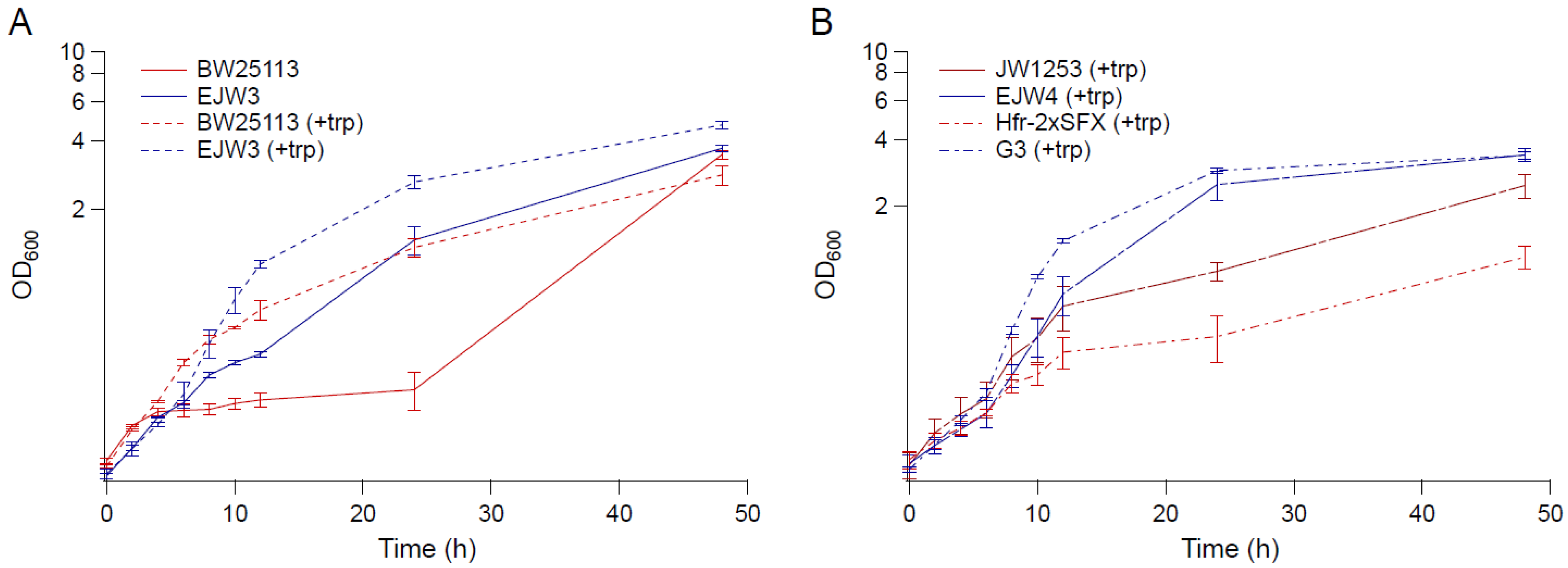
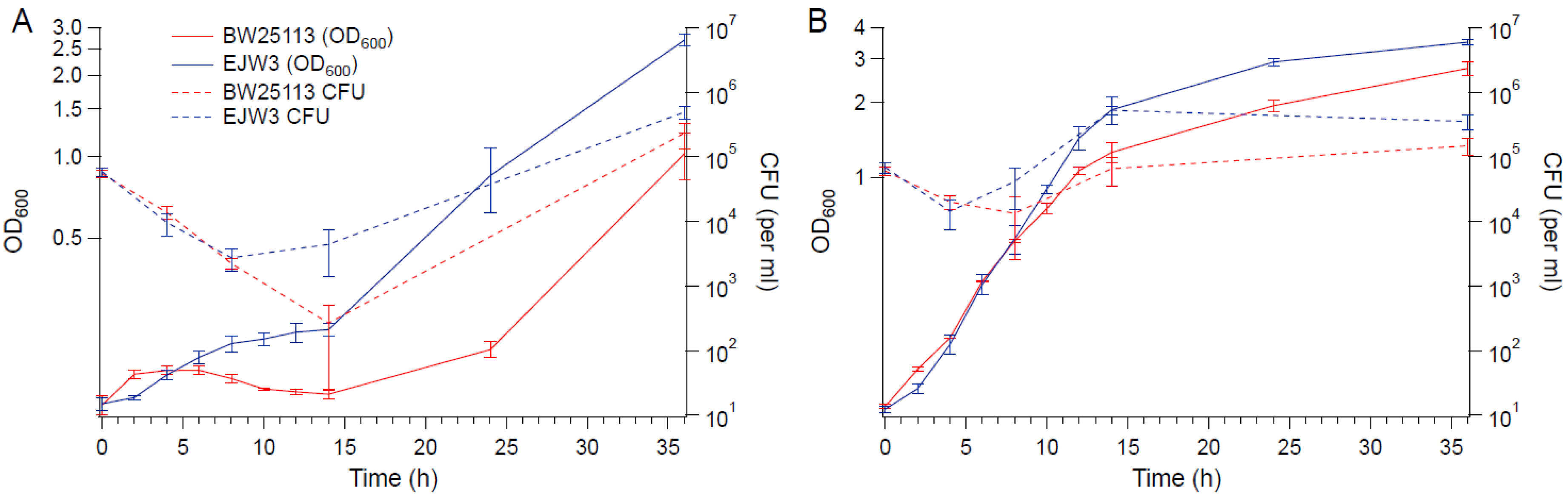
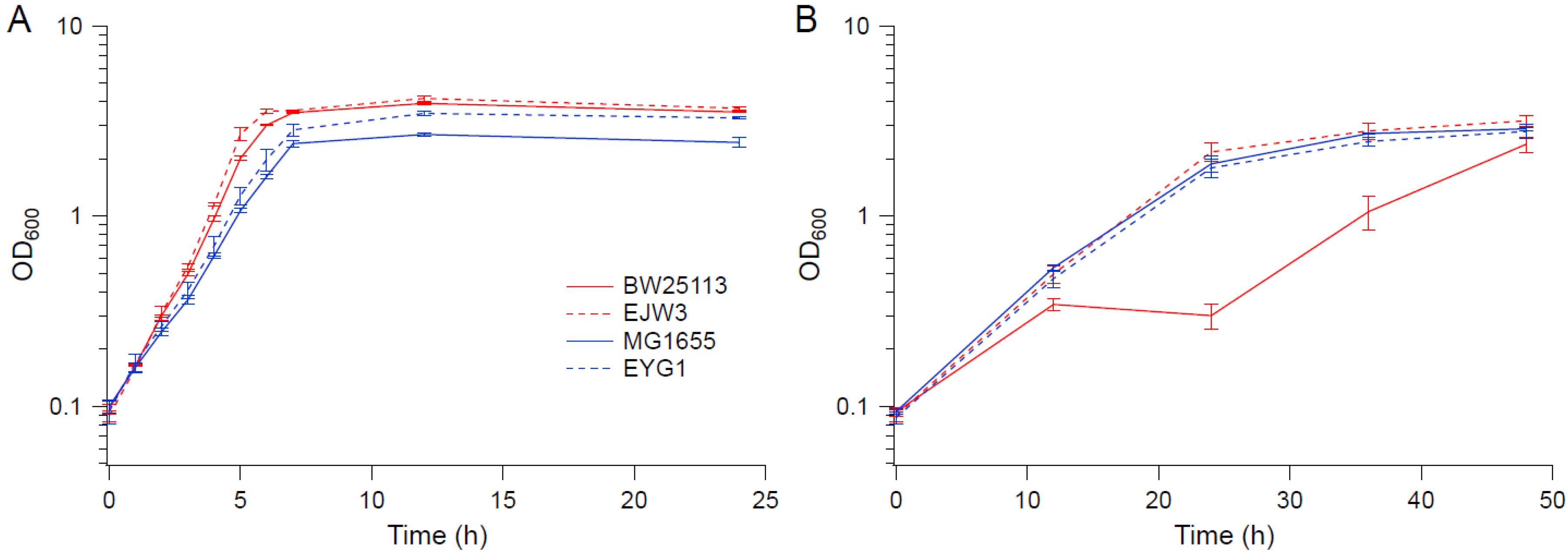
3.1. The rpoC K370_A396dup Mutation Confers Osmotic Tolerance in BW25113
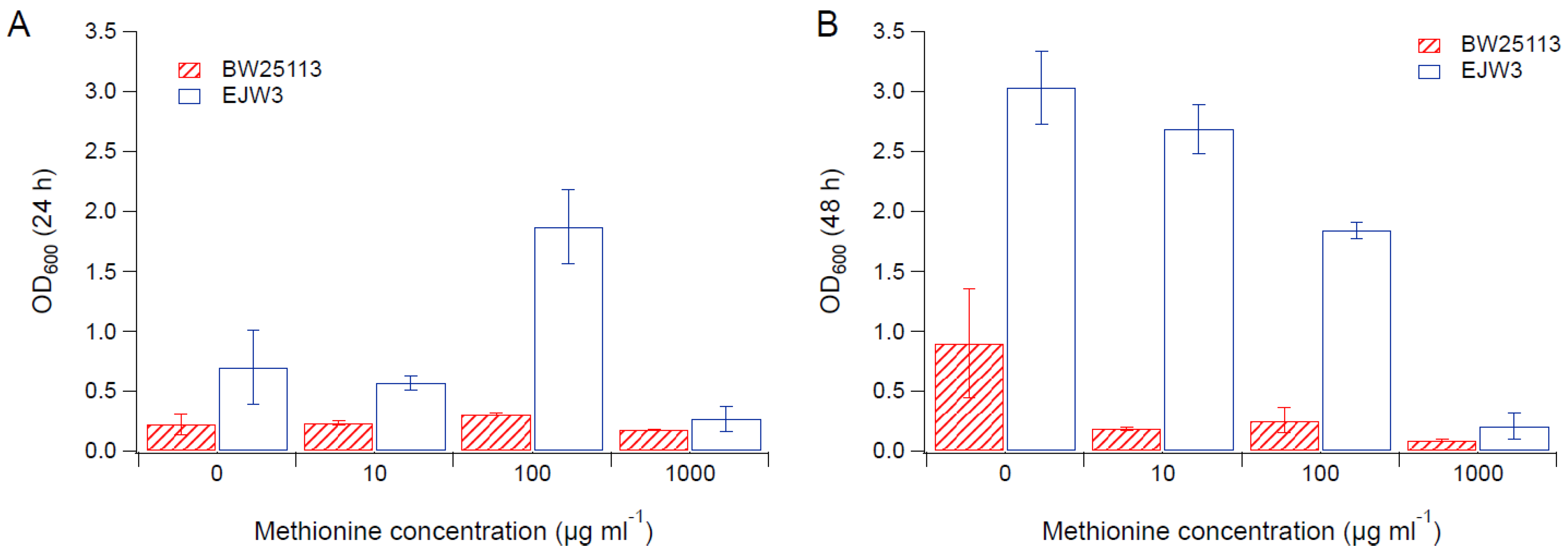
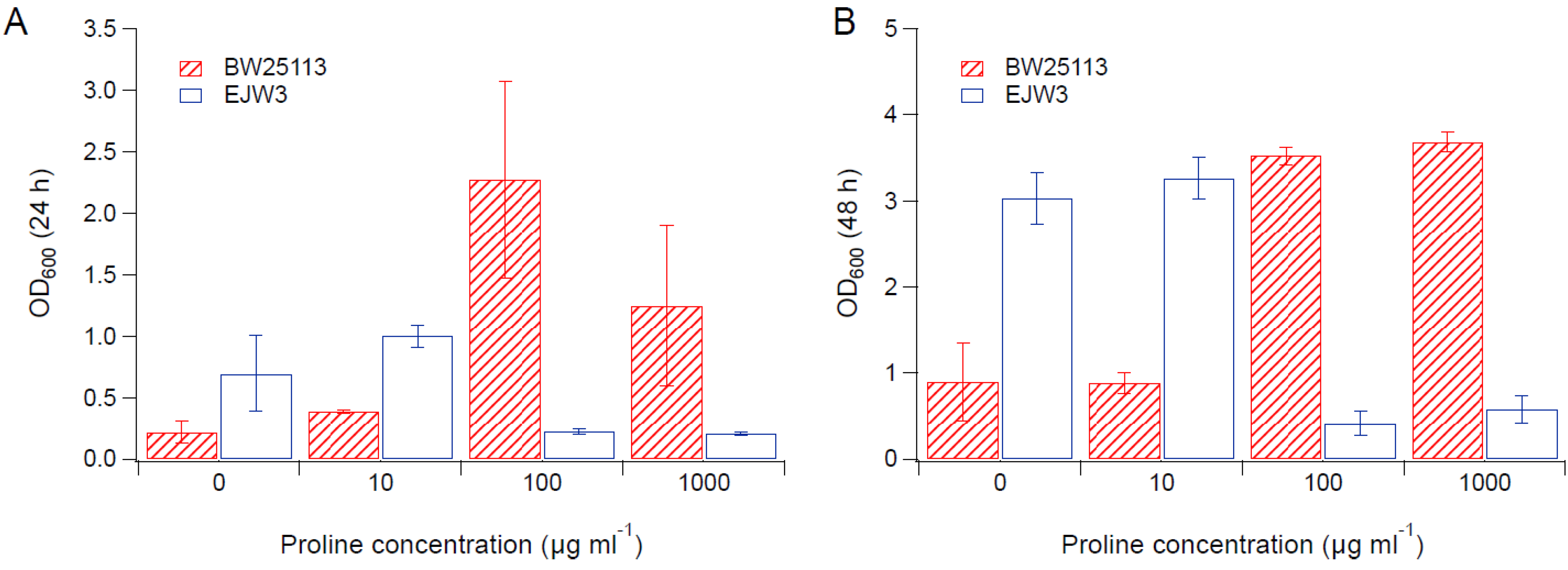
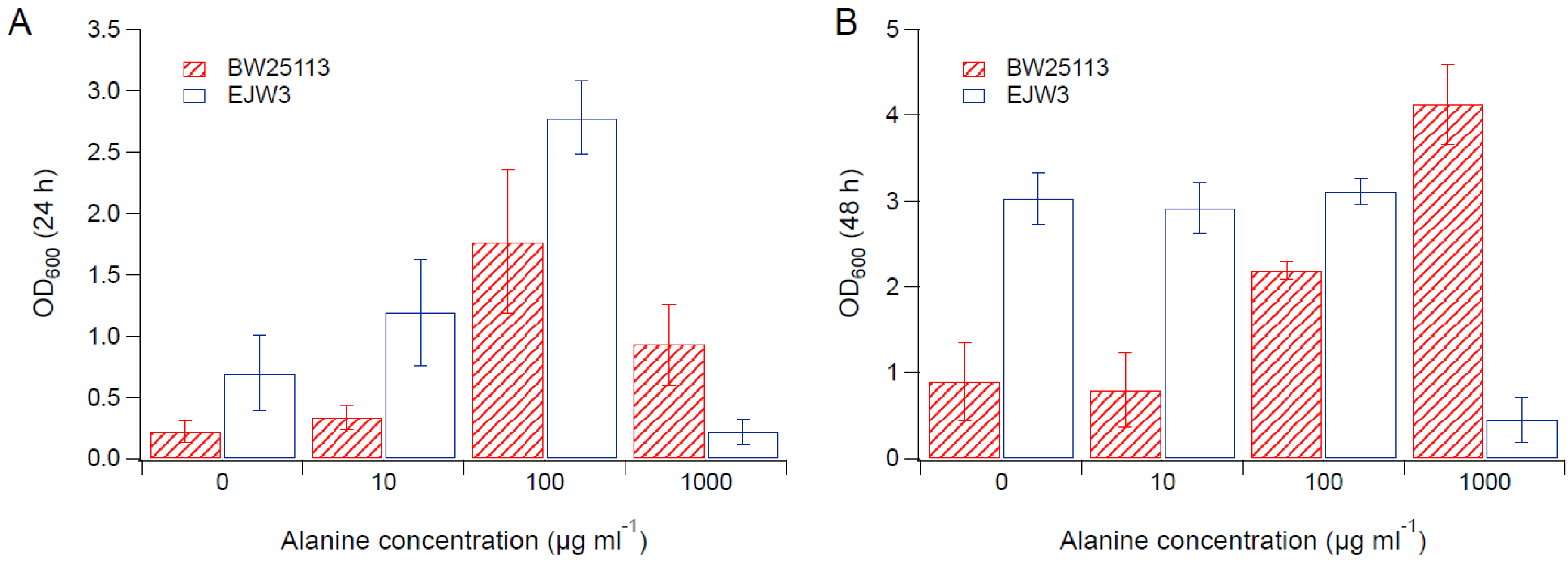
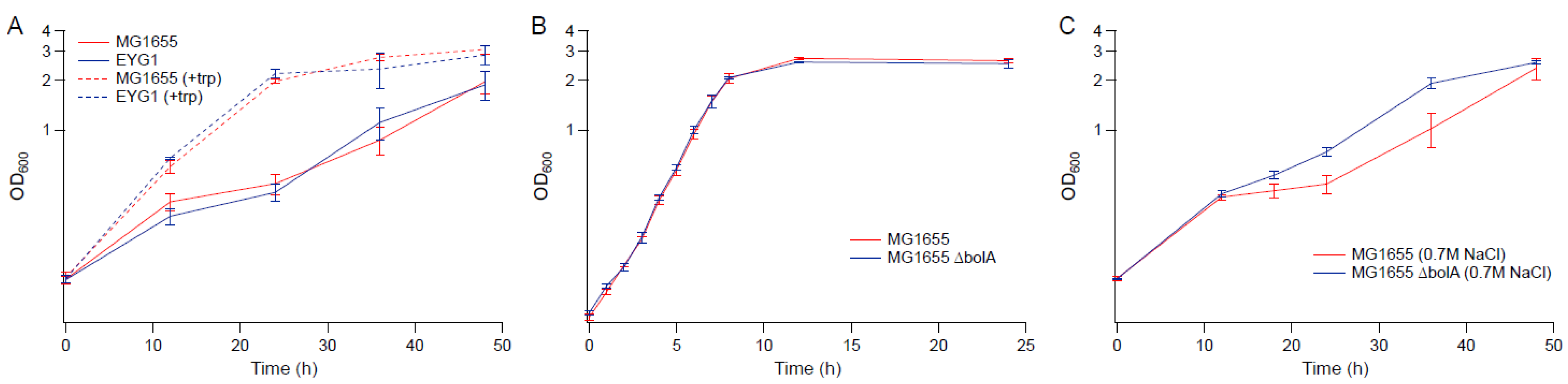
3.2. Effects of Amino Acid Supplementation
3.3. Metabolite Analysis
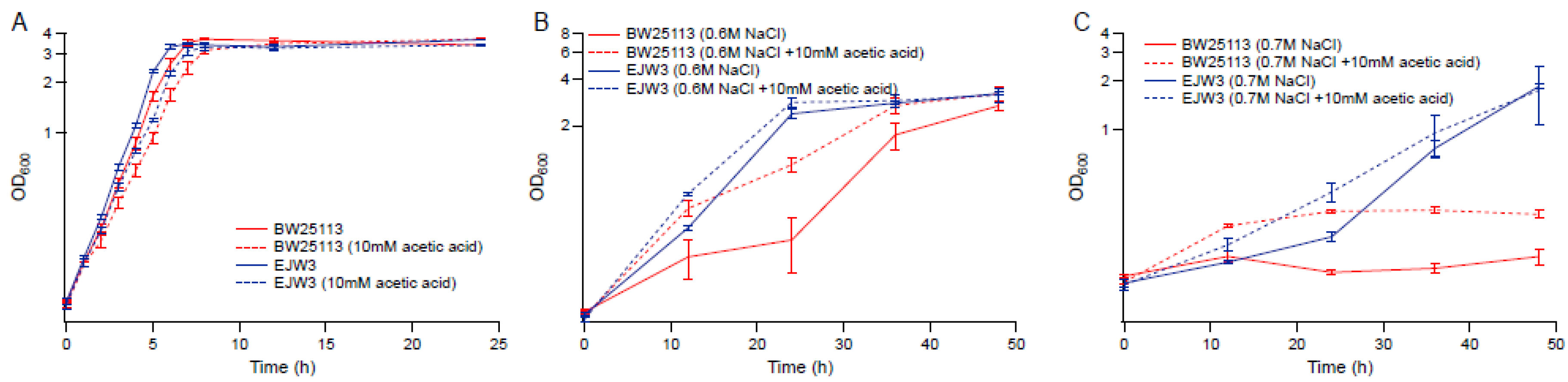
3.4. Effect of Acetic Acid on Osmotic Stress Tolerance
3.5. Cell Membrane Damage Analysis
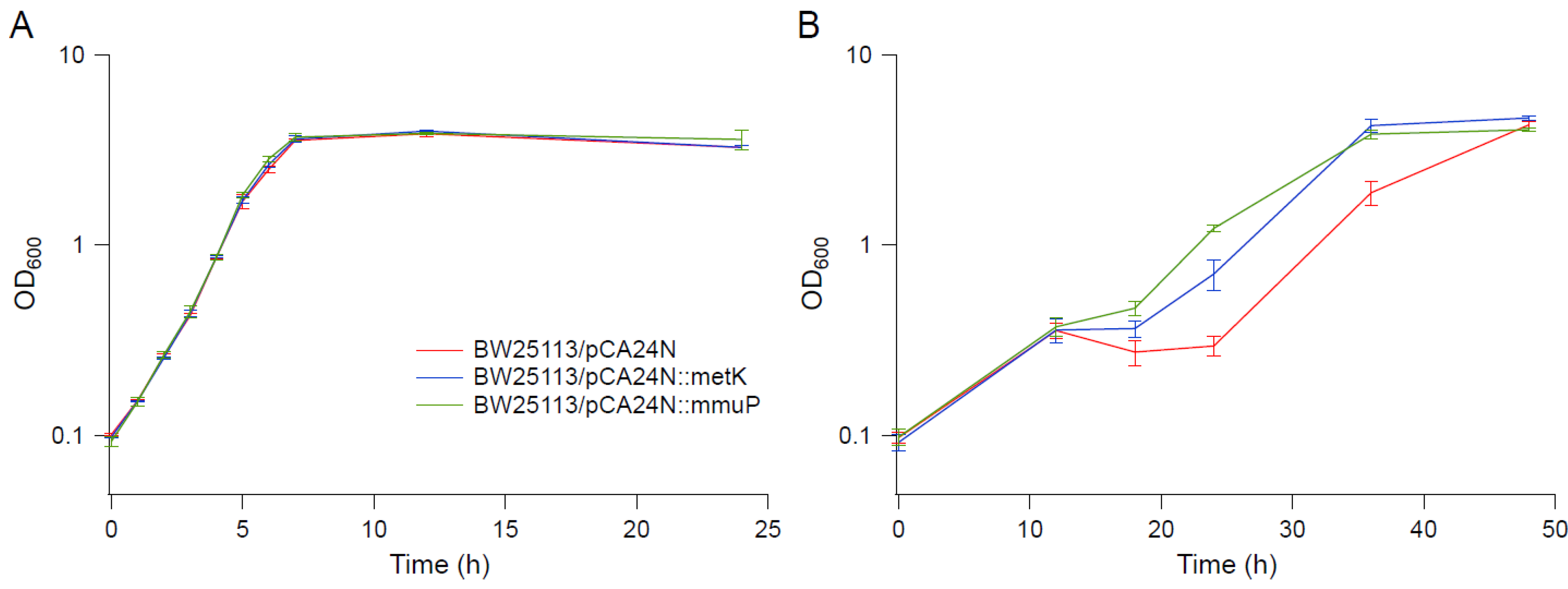
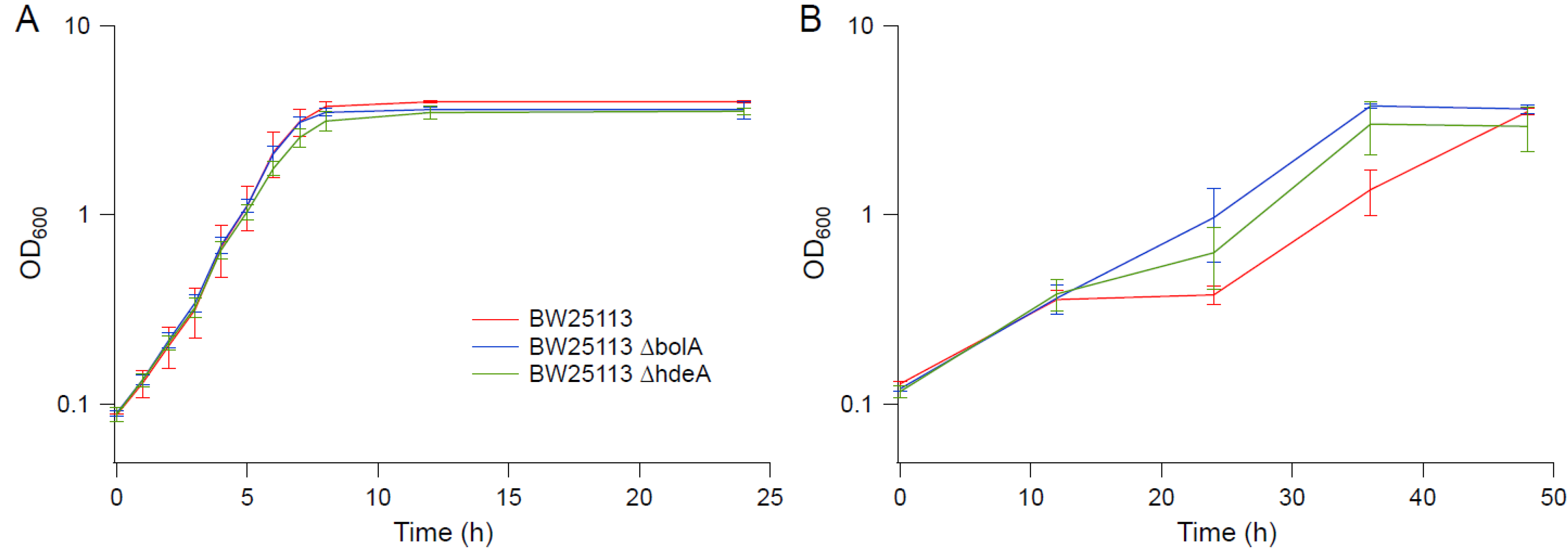
3.6. Transcriptional Profile Analysis
3.7. Impact of the rpoC Mutation in MG1655
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pleissner, D.; Neu, A.K.; Mehlmann, K.; Schneider, R.; Puerta-Quintero, G.I.; Venus, J. Fermentative lactic acid production from coffee pulp hydrolysate using Bacillus coagulans at laboratory and pilot scales. Bioresour. Technol. 2016, 218, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Saini, J.K.; Saini, R.; Tewari, L. Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: Concepts and recent developments. 3 Biotech 2015, 5, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Salvachua, D.; Smith, H.; St John, P.C.; Mohagheghi, A.; Peterson, D.J.; Black, B.A.; Dowe, N.; Beckham, G.T. Succinic acid production from lignocellulosic hydrolysate by Basfia succiniciproducens. Bioresour. Technol. 2016, 214, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lesnik, K.L.; Liu, H. Microbial conversion of waste glycerol from biodiesel production into value-added products. Energies 2013, 6, 4739–4768. [Google Scholar] [CrossRef]
- Przystalowska, H.; Zeyland, J.; Szymanowska-Powalowska, D.; Szalata, M.; Slomski, R.; Lipinski, D. 1,3-propanediol production by new recombinant Escherichia coli containing genes from pathogenic bacteria. Microbiol. Res. 2015, 171, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, J.S.; Zhang, Y.; Bates, D.M.; Keating, D.H.; Sato, T.K.; Ong, I.M.; Landick, R. Death by a thousand cuts: The challenges and diverse landscape of lignocellulosic hydrolysate inhibitors. Front. Microbiol 2014, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Rumbold, K.; van Buijsen, H.J.; Overkamp, K.M.; van Groenestijn, J.W.; Punt, P.J.; van der Werf, M.J. Microbial production host selection for converting second-generation feedstocks into bioproducts. Microb. Cell Fact. 2009, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Carlucci, A.F.; Pramer, D. An evaluation of factors affecting the survival of Escherichia coli in sea water. Ii. Salinity, ph, and nutrients. Appl. Microbiol. 1960, 8, 247–250. [Google Scholar] [PubMed]
- Pernetti, M.; Di Palma, L. Experimental evaluation of inhibition effects of saline wastewater on activated sludge. Environ. Technol. 2005, 26, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Delamarche, C.; Thomas, D.; Rolland, J.P.; Froger, A.; Gouranton, J.; Svelto, M.; Agre, P.; Calamita, G. Visualization of AqpZ-mediated water permeability in Escherichia coli by cryoelectron microscopy. J. Bacteriol. 1999, 181, 4193–4197. [Google Scholar] [PubMed]
- Laimins, L.A.; Rhoads, D.B.; Epstein, W. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 1981, 78, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Epstein, W. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Lett. 1986, 39, 73–78. [Google Scholar] [CrossRef]
- Landfald, B.; Strom, A.R. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 1986, 165, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Strøm, A.R.; Falkenberg, P.; Landfald, B. Genetics of osmoregulation in Escherichia coli: Uptake and biosynthesis of organic osmolytes. FEMS Microbiol. Lett. 1986, 39, 79–86. [Google Scholar] [CrossRef]
- Larsen, P.I.; Sydnes, L.K.; Landfald, B.; Strom, A.R. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: Betaines, glutamic acid, and trehalose. Arch. Microbiol. 1987, 147, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Perroud, B.; Le Rudulier, D. Glycine betaine transport in Escherichia coli: Osmotic modulation. J. Bacteriol. 1985, 161, 393–401. [Google Scholar] [PubMed]
- Sevin, D.C.; Sauer, U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 2014, 10, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Purvis, J.E.; Yomano, L.P.; Ingram, L.O. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl. Environ. Microbiol. 2005, 71, 3761–3769. [Google Scholar] [CrossRef] [PubMed]
- Rozwadowski, K.L.; Khachatourians, G.G.; Selvaraj, G. Choline oxidase, a catabolic enzyme in Arthrobacter pascens, facilitates adaptation to osmotic stress in Escherichia coli. J. Bacteriol. 1991, 173, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Wang, Y.G.; Yong, T.M.; She, Y.H.; Fu, F.L.; Li, W.C. Heterologous expression of betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus confers high salt and heat tolerance to Escherichia coli. Gene 2014, 549, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Xue, Y.; Song, Y.; Xian, M.; Yin, L.; Zhong, N.; Xia, G.; Ma, Y. Overexpression of AaPal, a peptidoglycan-associated lipoprotein from Alkalomonas amylolytica, improves salt and alkaline tolerance of Escherichia coli and Arabidopsis thaliana. Biotechnol. Lett. 2014, 36, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, J.; Zhou, Z.; Yan, Y.; Zhang, W.; Lu, W.; Ping, S.; Dai, Q.; Yuan, M.; Feng, B.; et al. IrrE, a global regulator of extreme radiation resistance in deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS ONE 2009, 4, e4422. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.I.; Lennen, R.M.; Herrgard, M.J.; Nielsen, A.T. Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress. Sci. Rep. 2015, 5, 17874. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.D.; Garcia, C.; Olson, M.; Callaway, E.; Kao, K.C. Evolved osmotolerant Escherichia coli mutants frequently exhibit defective N-acetylglucosamine catabolism and point mutations in cell shape-regulating protein MreB. Appl. Environ. Microbiol. 2014, 80, 3729–3740. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Lennox, E.S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1955, 1, 190–206. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; Kimball, E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 2007, 79, 6167–6173. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.W.; Brooks, A. Improved Amino Acid Methods Using Agilent ZORBAX Eclipse Plus C18 Columns for a Variety of Agilent LC Instrumentation and Separation Goals; Agilent Technologies: Santa Clara, CA, USA, 2010. [Google Scholar]
- Klotz, B.; Manas, P.; Mackey, B.M. The relationship between membrane damage, release of protein and loss of viability in Escherichia coli exposed to high hydrostatic pressure. Int. J. Food Microbiol. 2010, 137, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Pagan, R.; Mackey, B. Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: Differences between exponential- and stationary-phase cells and variation among strains. Appl. Environ. Microbiol. 2000, 66, 2829–2834. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [PubMed]
- Quackenbush, J. Microarray data normalization and transformation. Nat. Genet. 2002, 32, S496–S501. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Ni Bhriain, N.; Dorman, C.J.; Higgins, C.F. An overlap between osmotic and anaerobic stress responses: A potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol. Microbiol. 1989, 3, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Grothe, S.; Krogsrud, R.L.; McClellan, D.J.; Milner, J.L.; Wood, J.M. Proline transport and osmotic stress response in Escherichia coli K-12. J. Bacteriol. 1986, 166, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Akashi, H.; Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2002, 99, 3695–3700. [Google Scholar] [CrossRef] [PubMed]
- Glaser, H.U.; Thomas, D.; Gaxiola, R.; Montrichard, F.; Surdin-Kerjan, Y.; Serrano, R. Salt tolerance and methionine biosynthesis in Saccharomyces cerevisiae involve a putative phosphatase gene. EMBO J. 1993, 12, 3105–3110. [Google Scholar] [PubMed]
- Shahjee, H.M.; Banerjee, K.; Ahmad, F. Comparative analysis of naturally occurring l-amino acid osmolytes and their d-isomers on protection of Escherichia coli against environmental stresses. J. Biosci. 2002, 27, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Le Rudulier, D.; Bouillard, L. Glycine betaine, an osmotic effector in Klebsiella pneumoniae and other members of the Enterobacteriaceae. Appl. Environ. Microbiol. 1983, 46, 152–159. [Google Scholar] [PubMed]
- Csonka, L.N.; Gelvin, S.B.; Goodner, B.W.; Orser, C.S.; Siemieniak, D.; Slightom, J.L. Nucleotide sequence of a mutation in the proB gene of Escherichia coli that confers proline overproduction and enhanced tolerance to osmotic stress. Gene 1988, 64, 199–205. [Google Scholar] [CrossRef]
- Wilson, O.H.; Holden, J.T. Stimulation of arginine transport in osmotically shocked Escherichia coli W cells by purified arginine-binding protein fractions. J. Biol. Chem. 1969, 244, 2743–2749. [Google Scholar] [PubMed]
- Blum, J.J. Effects of osmotic stress on metabolism, shape, and amino acid content of Leishmania. Biol. Cell 1996, 87, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lim, H.C.; Hong, J. Acetic acid formation in Escherichia coli fermentation. Biotechnol. Bioeng. 1992, 39, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Castano-Cerezo, S.; Bernal, V.; Blanco-Catala, J.; Iborra, J.L.; Canovas, M. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol. Microbiol. 2011, 82, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Chapman, B.; Ross, T. Escherichia coli and Salmonella enterica are protected against acetic acid, but not hydrochloric acid, by hypertonicity. Appl. Environ. Microbiol. 2009, 75, 3605–3610. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.D.; Halweg-Edwards, A.L.; Erickson, K.E.; Choudhury, A.; Pines, G.; Gill, R.T. The resistome: A comprehensive database of Escherichia coli resistance phenotypes. ACS Synth. Biol. 2016, 5, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Fuhrer, T.; Zampieri, M.; Sevin, D.C.; Sauer, U.; Zamboni, N. Genomewide landscape of gene-metabolome associations in Escherichia coli. Mol. Syst. Biol. 2017, 13, 907. [Google Scholar] [CrossRef] [PubMed]
- Smelt, J.P.P.M.; Rijke, A.G.F.; Hayhurst, A. Possible mechanism of high pressure inactivation of microorganisms. High Press. Res. 1994, 12, 199–203. [Google Scholar] [CrossRef]
- Benito, A.; Ventoura, G.; Casadei, M.; Robinson, T.; Mackey, B. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 1999, 65, 1564–1569. [Google Scholar] [PubMed]
- Arthur, T.M.; Burgess, R.R. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta’ subunit. J. Biol. Chem. 1998, 273, 31381–31387. [Google Scholar] [CrossRef] [PubMed]
- Brodolin, K.; Mustaev, A.; Severinov, K.; Nikiforov, V. Identification of RNA polymerase beta’ subunit segment contacting the melted region of the lacUV5 promoter. J. Biol. Chem. 2000, 275, 3661–3666. [Google Scholar] [CrossRef] [PubMed]
- Zaychikov, E.; Martin, E.; Denissova, L.; Kozlov, M.; Markovtsov, V.; Kashlev, M.; Heumann, H.; Nikiforov, V.; Goldfarb, A.; Mustaev, A. Mapping of catalytic residues in the RNA polymerase active center. Science 1996, 273, 107–109. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.S. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. J. Biol. Chem. 2013, 288, 9126–9134. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Wang, D.; Ebright, Y.W.; Korlann, Y.; Kortkhonjia, E.; Kim, T.; Chowdhury, S.; Wigneshweraraj, S.; Irschik, H.; Jansen, R.; et al. Opening and closing of the bacterial RNA polymerase clamp. Science 2012, 337, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Zaychikov, E.; Denissova, L.; Meier, T.; Gotte, M.; Heumann, H. Influence of Mg2+ and temperature on formation of the transcription bubble. J. Biol. Chem. 1997, 272, 2259–2267. [Google Scholar] [CrossRef] [PubMed]
- Vrentas, C.E.; Gaal, T.; Ross, W.; Ebright, R.H.; Gourse, R.L. Response of RNA polymerase to ppGpp: Requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005, 19, 2378–2387. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Wang, Y.; Steitz, T.A. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell 2013, 50, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Ross, W.; Vrentas, C.E.; Sanchez-Vazquez, P.; Gaal, T.; Gourse, R.L. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 2013, 50, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, L.U.; Farewell, A.; Nystrom, T. Ppgpp: A global regulator in Escherichia coli. Trends Microbiol. 2005, 13, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Markham, G.D.; Hafner, E.W.; Tabor, C.W.; Tabor, H. S-adenosylmethionine synthetase from Escherichia coli. J. Biol. Chem. 1980, 255, 9082–9092. [Google Scholar] [PubMed]
- Thomas, D.; Cherest, H.; Surdin-Kerjan, Y. Identification of the structural gene for glucose-6-phosphate dehydrogenase in yeast. Inactivation leads to a nutritional requirement for organic sulfur. EMBO J. 1991, 10, 547–553. [Google Scholar] [PubMed]
- Sekowska, A.; Kung, H.F.; Danchin, A. Sulfur metabolism in Escherichia coli and related bacteria: Facts and fiction. J. Mol. Microbiol. Biotechnol. 2000, 2, 145–177. [Google Scholar] [PubMed]
- Chen, D.; Ma, H.; Hong, H.; Koh, S.S.; Huang, S.M.; Schurter, B.T.; Aswad, D.W.; Stallcup, M.R. Regulation of transcription by a protein methyltransferase. Science 1999, 284, 2174–2177. [Google Scholar] [CrossRef] [PubMed]
- Augustus, A.M.; Spicer, L.D. The MetJ regulon in gammaproteobacteria determined by comparative genomics methods. BMC Genom. 2011, 12, 558. [Google Scholar] [CrossRef] [PubMed]
- Thanbichler, M.; Neuhierl, B.; Bock, A. S-methylmethionine metabolism in Escherichia coli. J. Bacteriol. 1999, 181, 662–665. [Google Scholar] [PubMed]
- Szego, D.; Kósa, E.; Horváth, E. Role of S-methylmethionine in the plant metabolism. Acta Agron. Hung. 2007, 55. [Google Scholar] [CrossRef]
- Santos, J.M.; Freire, P.; Vicente, M.; Arraiano, C.M. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol. Microbiol. 1999, 32, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Gajiwala, K.S.; Burley, S.K. HdeA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria1. J. Mol. Biol. 2000, 295, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Church, G.M. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 2003, 48, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Bittner, A.N.; Wang, J.D. Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 2015, 24, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Srivatsan, A.; Wang, J.D. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 2008, 11, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Tarusawa, T.; Ito, S.; Goto, S.; Ushida, C.; Muto, A.; Himeno, H. (p)ppGpp-dependent and -independent pathways for salt tolerance in Escherichia coli. J. Biochem. 2016, 160, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Maitra, A.; Shulgina, I.; Hernandez, V.J. Conversion of active promoter-RNA polymerase complexes into inactive promoter bound complexes in E. coli by the transcription effector, ppGpp. Mol. Cell 2005, 17, 817–829. [Google Scholar] [CrossRef] [PubMed]

| Strains | Description/Genotype | Source |
|---|---|---|
| BW25113 | F-, Δ(araD-araB)567, lacZ4787(del)::rrnB-3, λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514 | CGSC |
| MG655 | F-, λ-, rph-1 | CGSC |
| Hfr-2×SFX- | BW25113 ΔmbhA::oriT, ΔhyfC::oriT, trp::F[ΔtraST], (genR), parental strain of G3 | [24] |
| G3 | Evolved mutant of Hfr-2×SFX- containing rpoC K370_A396dup mutation | [24] |
| JW3929 | BW25113 ΔargE744::kan | [25] |
| JW1253 | BW25113 ΔtrpB769::kan | [25] |
| EJW1 | G3 ΔargE744::kan | This study |
| EJW2 | BW25113 ΔargE744::kan, rpoC K370_A396dup | This study |
| EJW3 | BW25113 rpoC K370_A396dup | This study |
| EJW4 | BW25113 rpoC K370_A396dup, ΔtrpB769::kan | This study |
| EYG1 | MG1655 rpoC K370_A396dup | This study |
| Strains | 0 h | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| BW25113 | − | + | + | + |
| EJW3 | − | + | +++ | ++++ |
| JW1253 * | − | + | ++ | + |
| EJW4 * | − | ++ | ++++ | ++++ |
| Hfr-2×SFX- * | − | + | + | + |
| G3 * | − | +++ | ++++ | +++ |
| Strains | 0 h | 24 h | 48 h | 72 h |
|---|---|---|---|---|
| BW25113 | − | + | + | + |
| EJW3 | − | + | +++ | ++++ |
| JW1253 * | − | + | ++ | + |
| EJW4 * | − | ++ | ++++ | ++++ |
| Hfr-2×SFX- * | − | + | + | + |
| G3 * | − | +++ | ++++ | +++ |
| Metabolite | BW25113 | EJW3 | p-Value | |
|---|---|---|---|---|
| Intracellular | Trehalose (µg mL−1 OD600−1) | - | - | - |
| Acetic acid (µg mL−1 OD600−1) | 650 ± 90 | 390 ± 80 | 0.000 * | |
| Glu (µg mL−1 OD600−1) | 187.72 ± 35.94 | 212.37 ± 27.2 | 0.210 | |
| Ala (µg mL−1 OD600−1) | 81.18 ± 16.55 | 83.71 ± 3.15 | 0.721 | |
| Arg (µg mL−1 OD600−1) | - | 4.78 ± 7.41 | 0.145 | |
| Pro (µg mL−1 OD600−1) | 61.64 ± 6.12 | 75.06 ± 1.95 | 0.000 * | |
| Extracellular | Trehalose (µg mL−1 OD600−1) | - | - | - |
| Acetic acid (µg mL−1 OD600−1) | 280 ± 20 | 370 ± 10 | 0.000 * | |
| Glu (µg mL−1 OD600−1) | - | 6.42 ± 12.41 | 0.234 | |
| Ala (µg mL−1 OD600−1) | 10.86 ± 6.33 | 1.93 ± 4.72 | 0.020 * | |
| Arg (µg mL−1 OD600−1) | 2.95 ± 7.21 | - | 0.341 | |
| Pro (µg mL−1 OD600−1) | 516.13 ± 35.09 | 776.02 ± 117.02 | 0.000 * |
| Metabolite | BW25113 | EJW3 | p-Value | |
|---|---|---|---|---|
| Intracellular | Trehalose (µg mL−1 OD600−1) | 1190 ± 190 | 1630 ± 220 | 0.005 * |
| Acetic acid (µg mL−1 OD600−1) | 270 ± 140 | 370 ± 50 | 0.137 | |
| Glu (µg mL−1 OD600−1) | 565.50 ± 44.39 | 698.75 ± 82.47 | 0.006 * | |
| Ala (µg mL−1 OD600−1) | 38.49 ± 9.03 | 71.58 ± 13.51 | 0.001 * | |
| Arg (µg mL−1 OD600−1) | 28.79 ± 2.73 | 44.87 ± 6.42 | 0.000 * | |
| Pro (µg mL−1 OD600−1) | 94.31 ± 6.14 | 107.31 ± 18.37 | 0.131 | |
| Extracellular | Trehalose (µg mL−1 OD600−1) | - | - | - |
| Acetic acid (µg mL−1 OD600−1) | 260 ± 20 | 360 ± 20 | 0.000 * | |
| Glu (µg mL−1 OD600−1) | 234.82 ± 15.44 | 88.05 ± 19.38 | 0.000 * | |
| Ala (µg mL−1 OD600−1) | - | - | - | |
| Arg (µg mL−1 OD600−1) | - | 7.90 ± 12.34 | 0.148 | |
| Pro (µg mL−1 OD600−1) | 747.75 ± 120.88 | 728.90 ± 156.43 | 0.820 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Winkler, J.; Kao, K.C. Insights on Osmotic Tolerance Mechanisms in Escherichia coli Gained from an rpoC Mutation. Bioengineering 2017, 4, 61. https://doi.org/10.3390/bioengineering4030061
Guo Y, Winkler J, Kao KC. Insights on Osmotic Tolerance Mechanisms in Escherichia coli Gained from an rpoC Mutation. Bioengineering. 2017; 4(3):61. https://doi.org/10.3390/bioengineering4030061
Chicago/Turabian StyleGuo, Yuqi, James Winkler, and Katy C. Kao. 2017. "Insights on Osmotic Tolerance Mechanisms in Escherichia coli Gained from an rpoC Mutation" Bioengineering 4, no. 3: 61. https://doi.org/10.3390/bioengineering4030061





