Development of Self-Assembled Nanoribbon Bound Peptide-Polyaniline Composite Scaffolds and Their Interactions with Neural Cortical Cells
Abstract
:1. Introduction
2. Materials and Methods
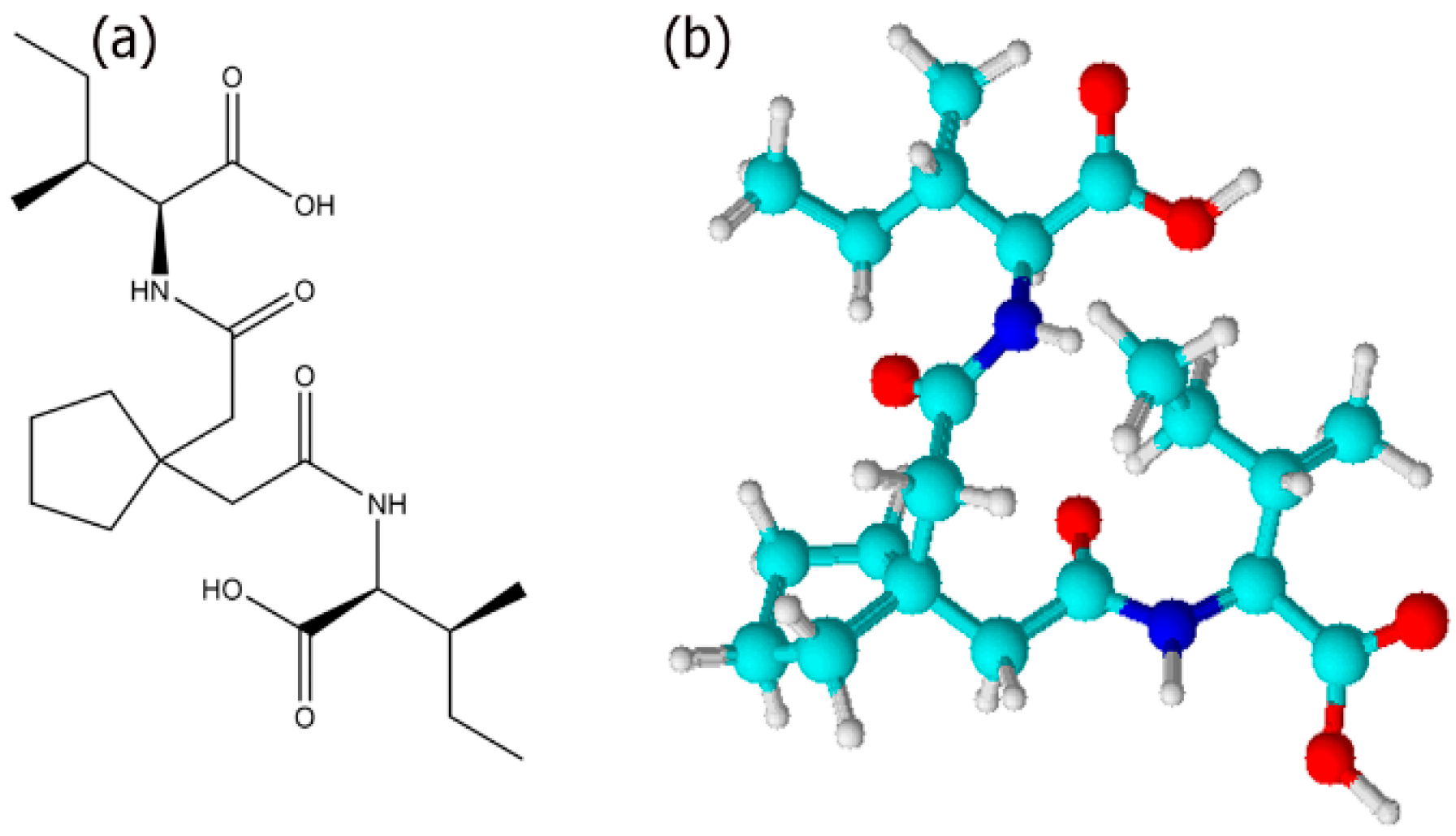
2.1. Synthesis of Ile-TMG-Ile
2.2. Self-Assembly of Ile-TMG-Ile Template
2.3. Preparation of Scaffold
2.4. Binding Efficiency of Laminin, Artemin and Type IV Collagen on the Assemblies
2.5. In Vitro Biodegradability Studies
2.6. Cell Studies
2.7. Electrochemical Studies
2.8. Characterization
2.8.1. Fourier Transform Infrared (FTIR) Spectroscopy
2.8.2. Scanning Electron Microscopy (SEM)
2.8.3. Transmission Electron Microscopy (TEM)
2.8.4. Dynamic Light Scattering (DLS)
2.8.5. Atomic Force Microscopy (AFM)
2.8.6. Differential Scanning Calorimetry (DSC)
2.8.7. Fluorescence Microscopy
2.8.8. UV-Vis Spectroscopy
2.9. Statistical Analysis
3. Results and Discussion
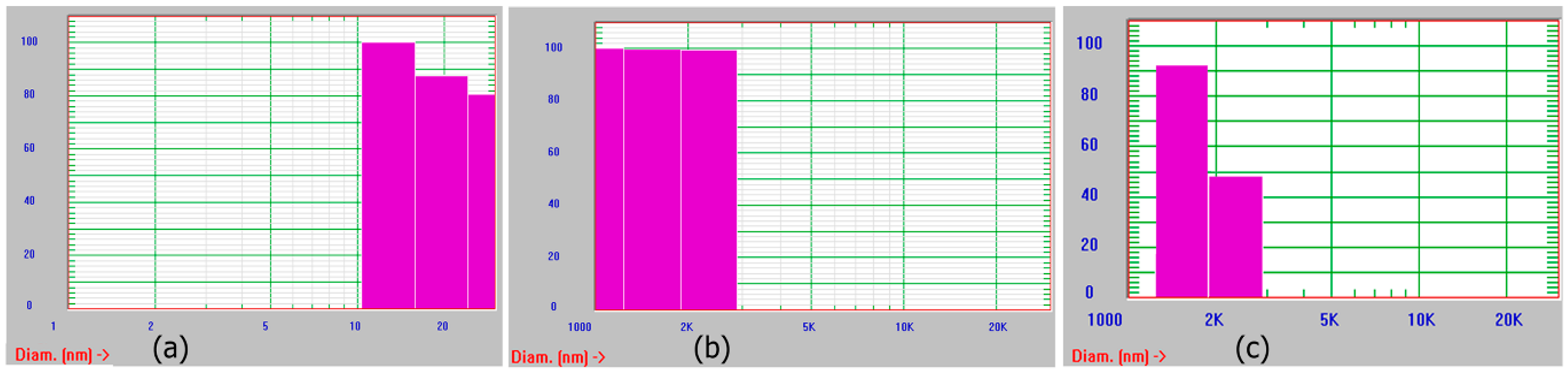
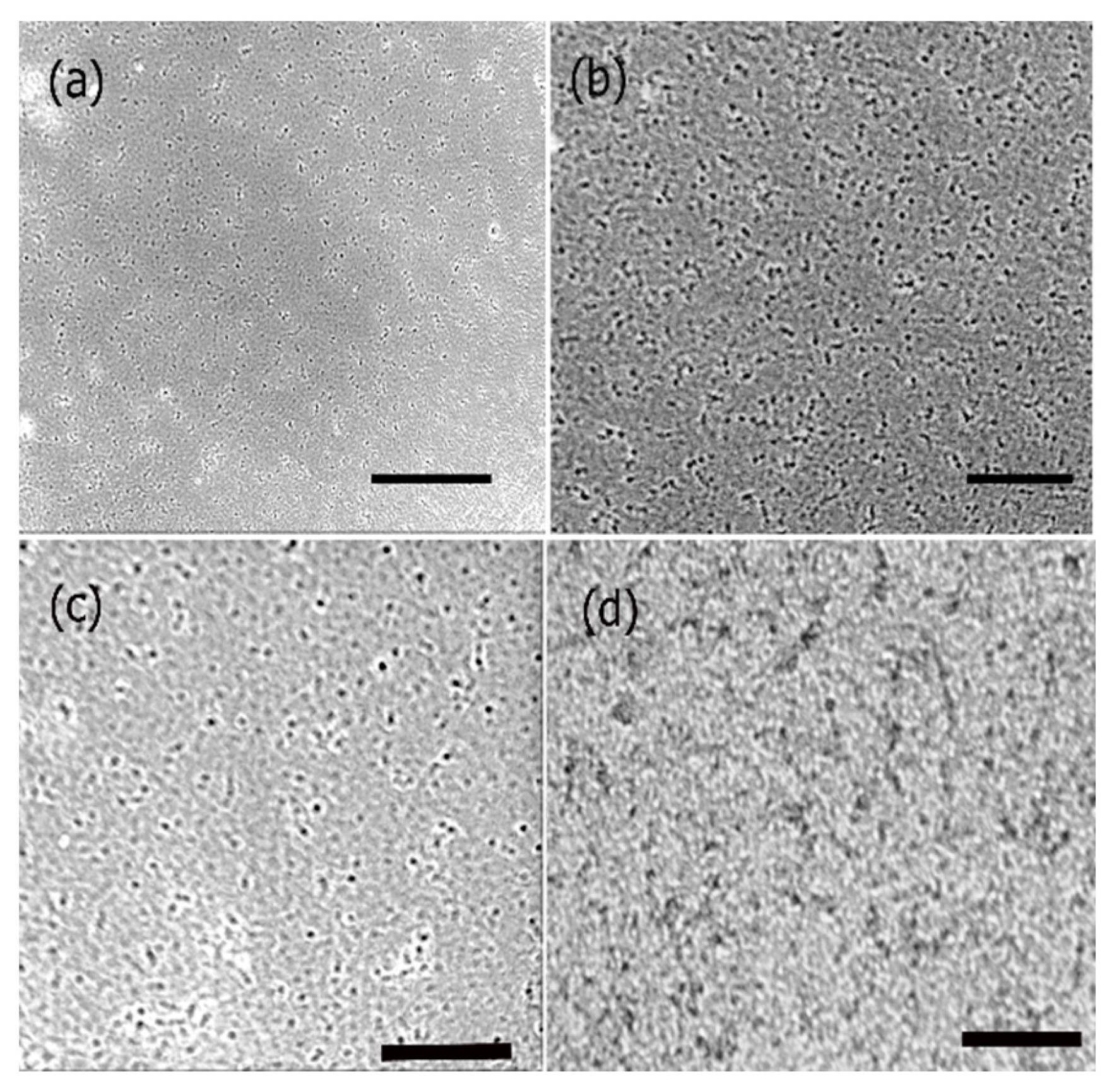
3.1. Self-Assembly of Ile-TMG-Ile
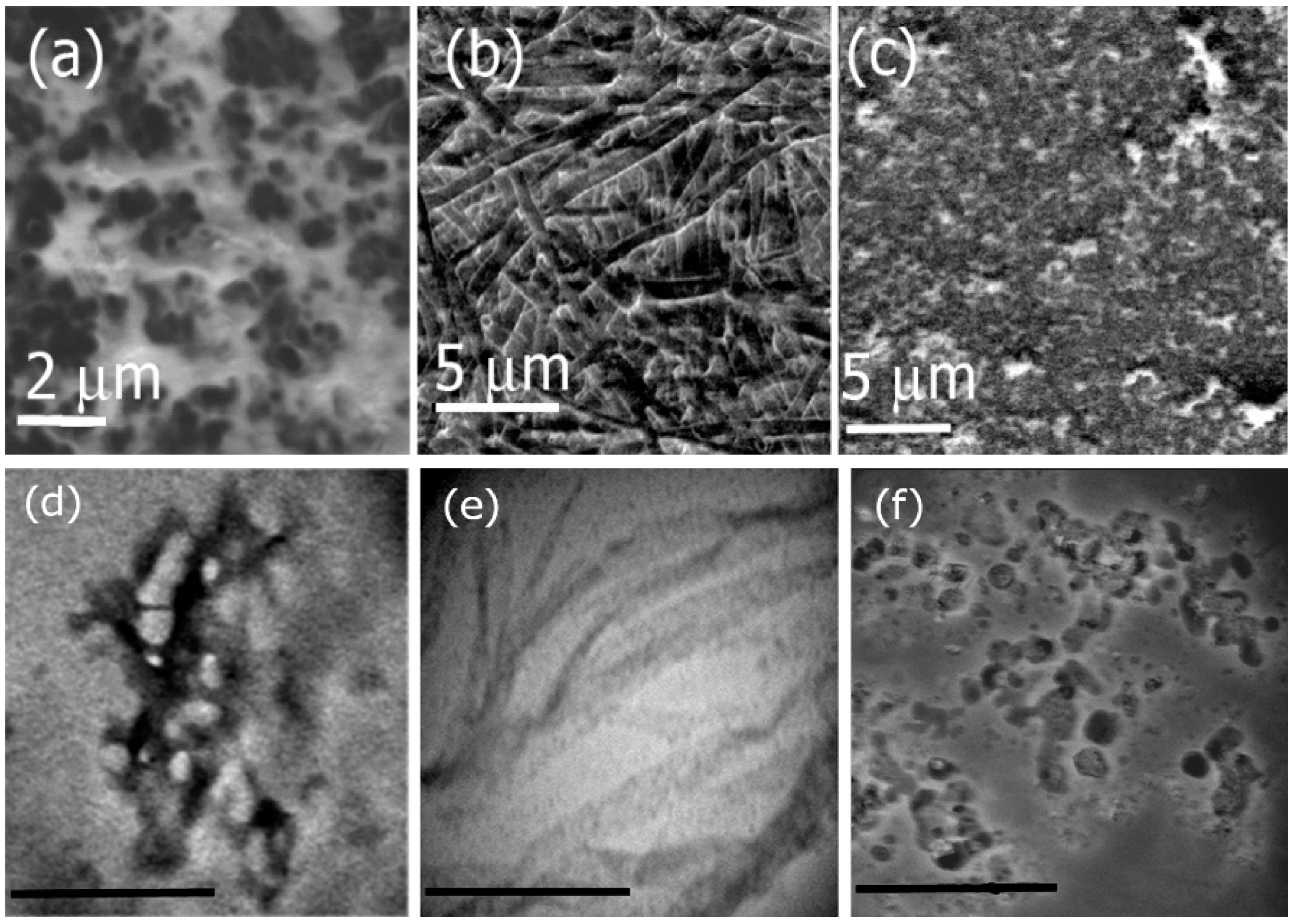
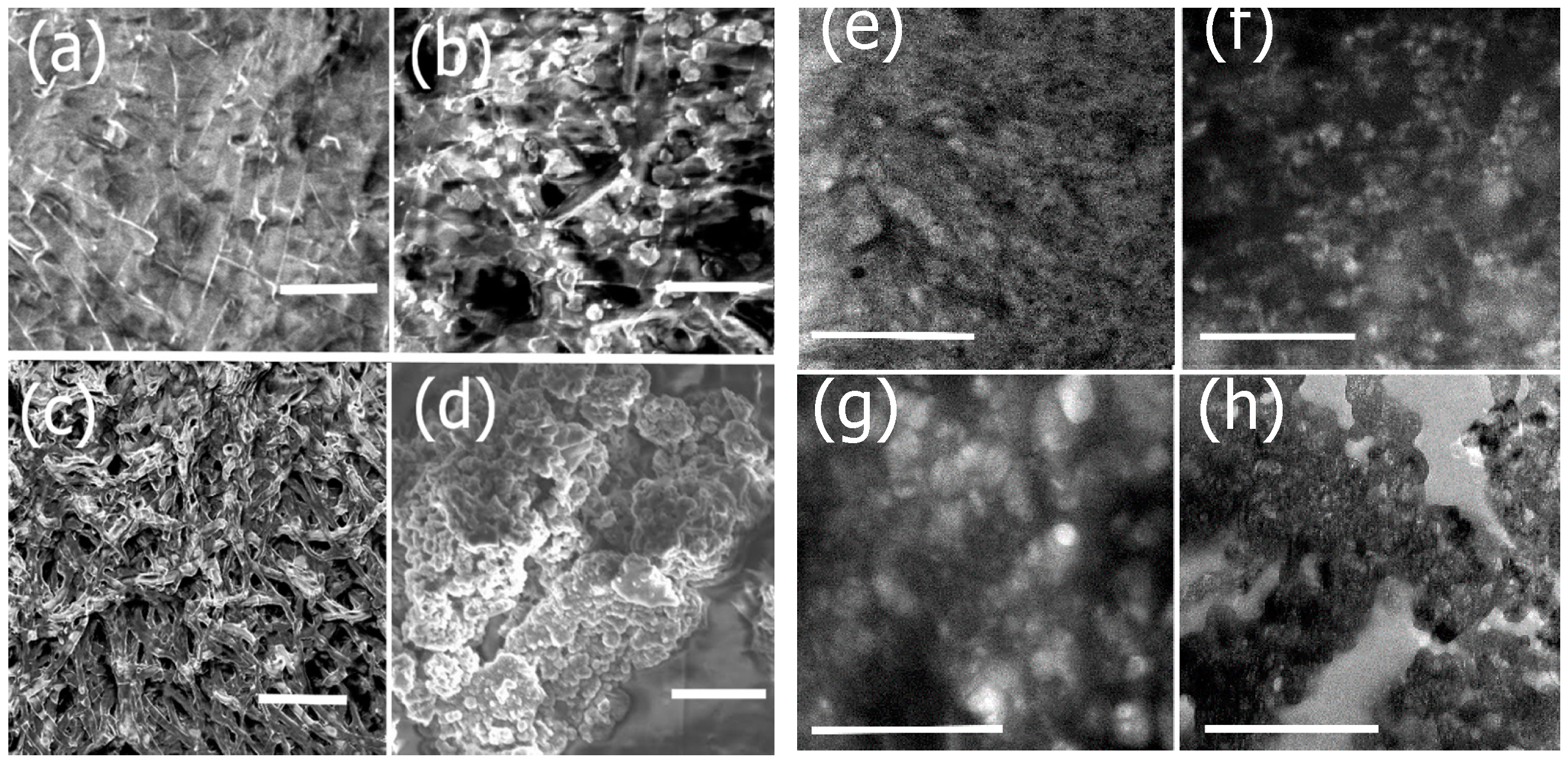
3.2. Functionalization of Ile-TMG-Ile and Preparation of Scaffold
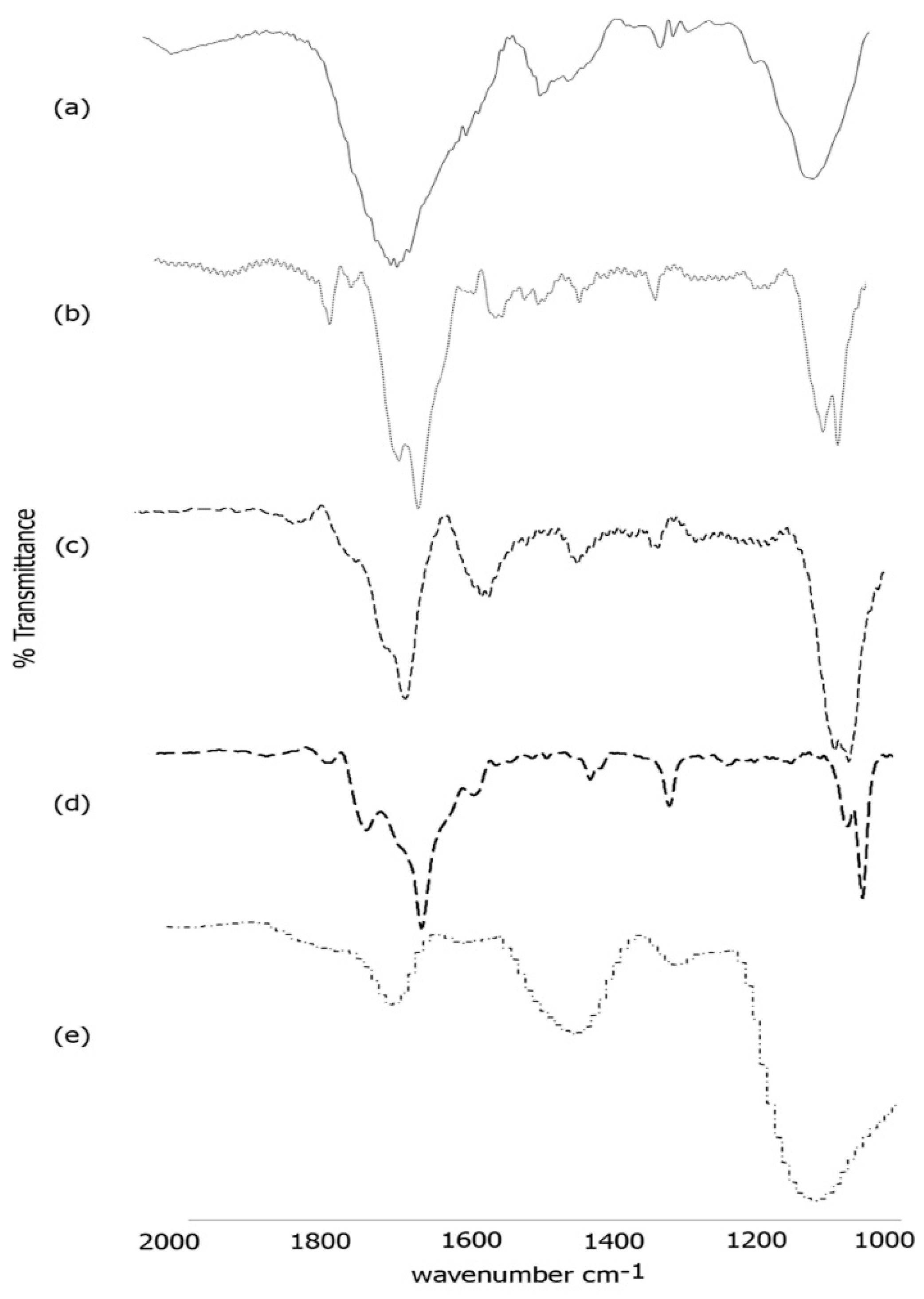
3.3. FTIR Spectroscopy
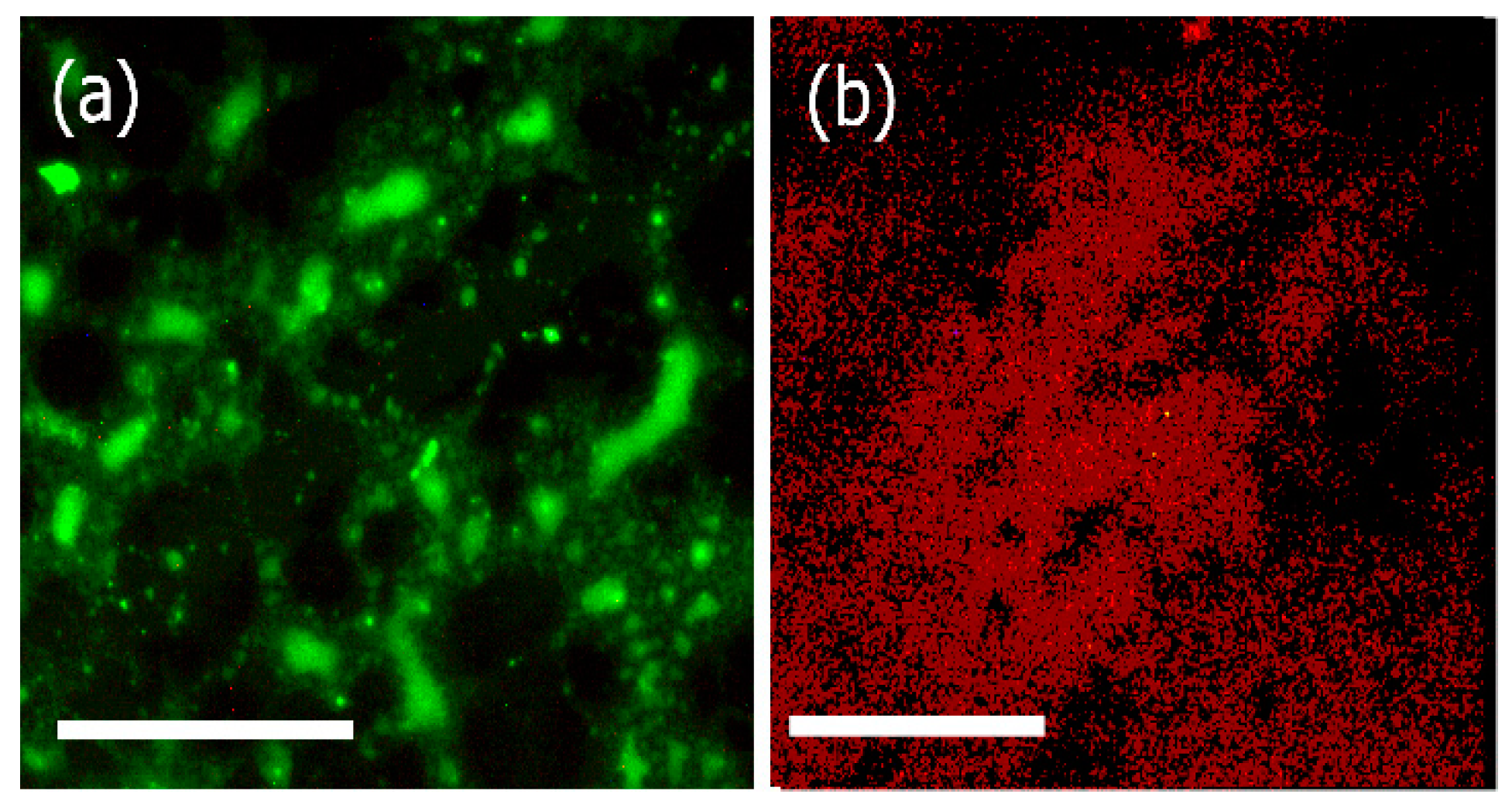
3.4. Fluorescence Microscopy
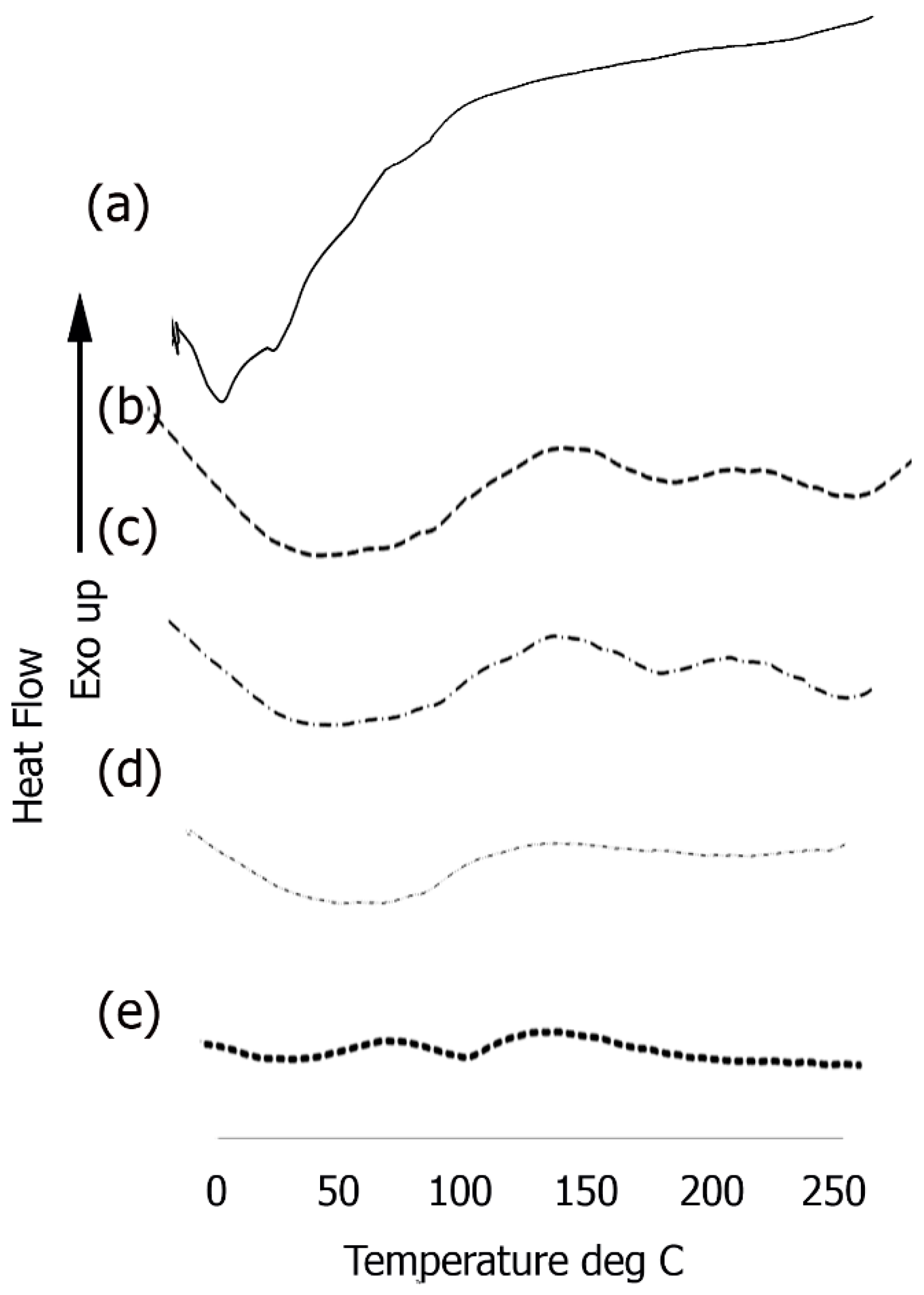
3.5. Differential Scanning Calorimetry
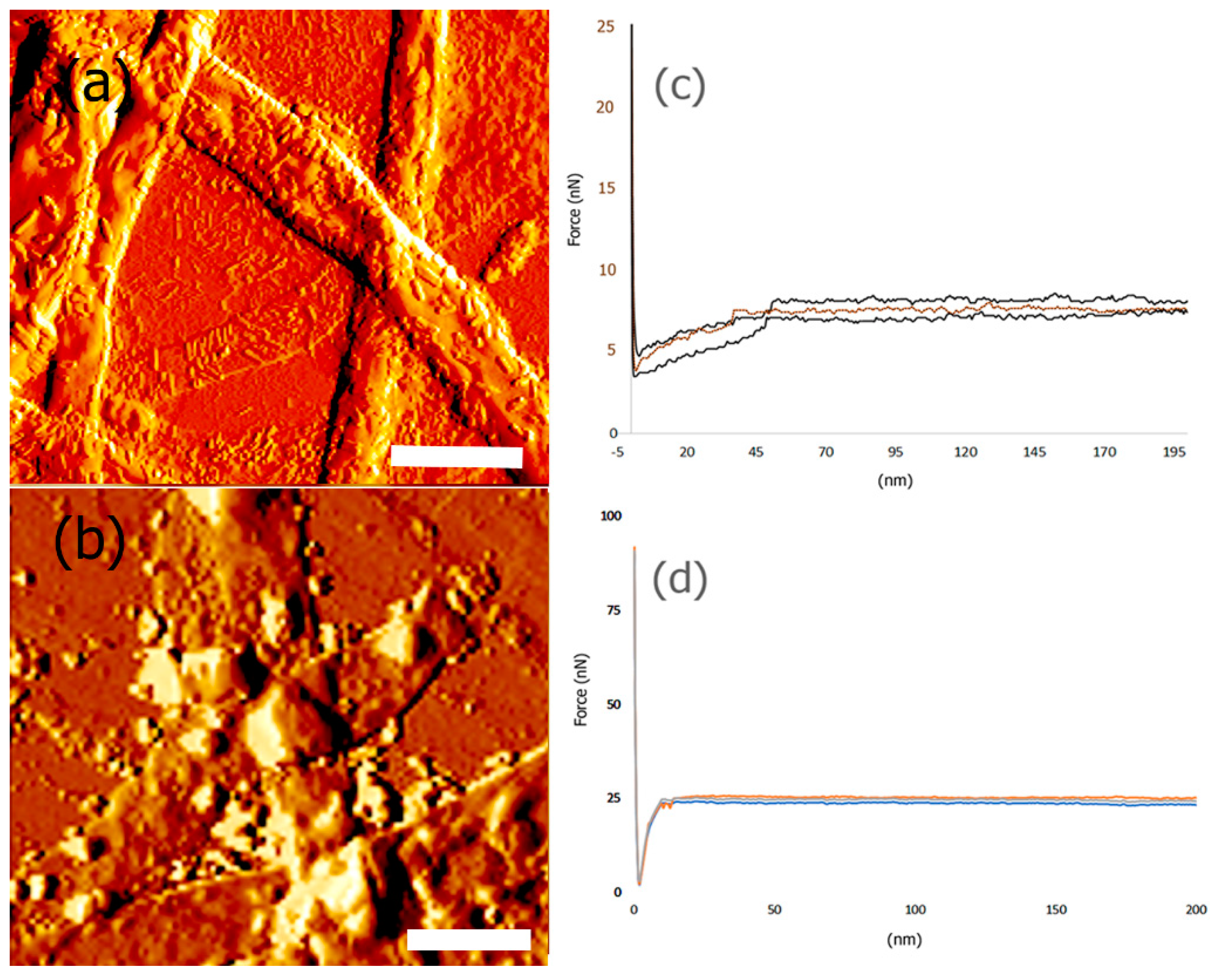
3.6. Mechanical and Surface Properties
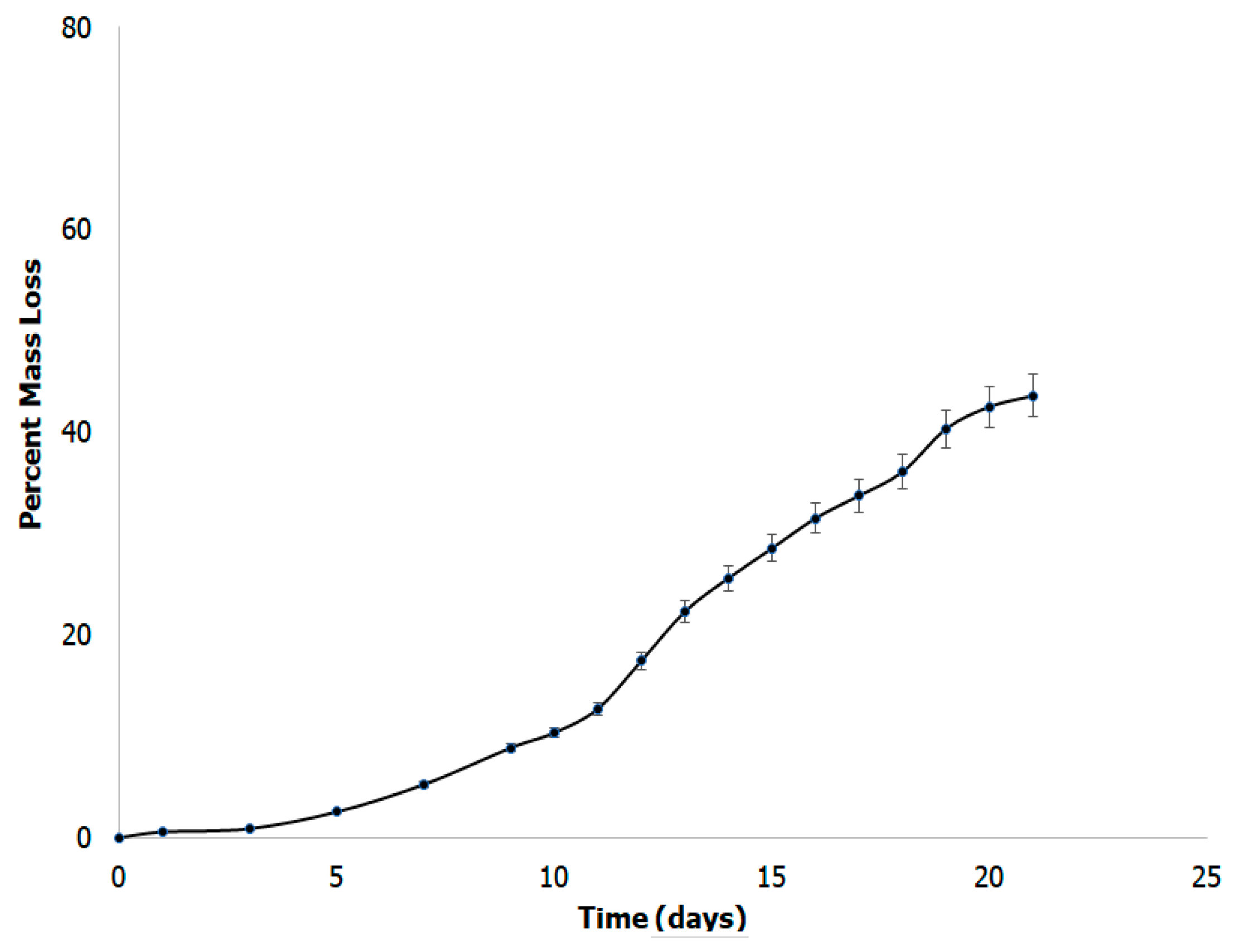
3.7. In Vitro Biodegradability Studies
3.8. Cell Studies
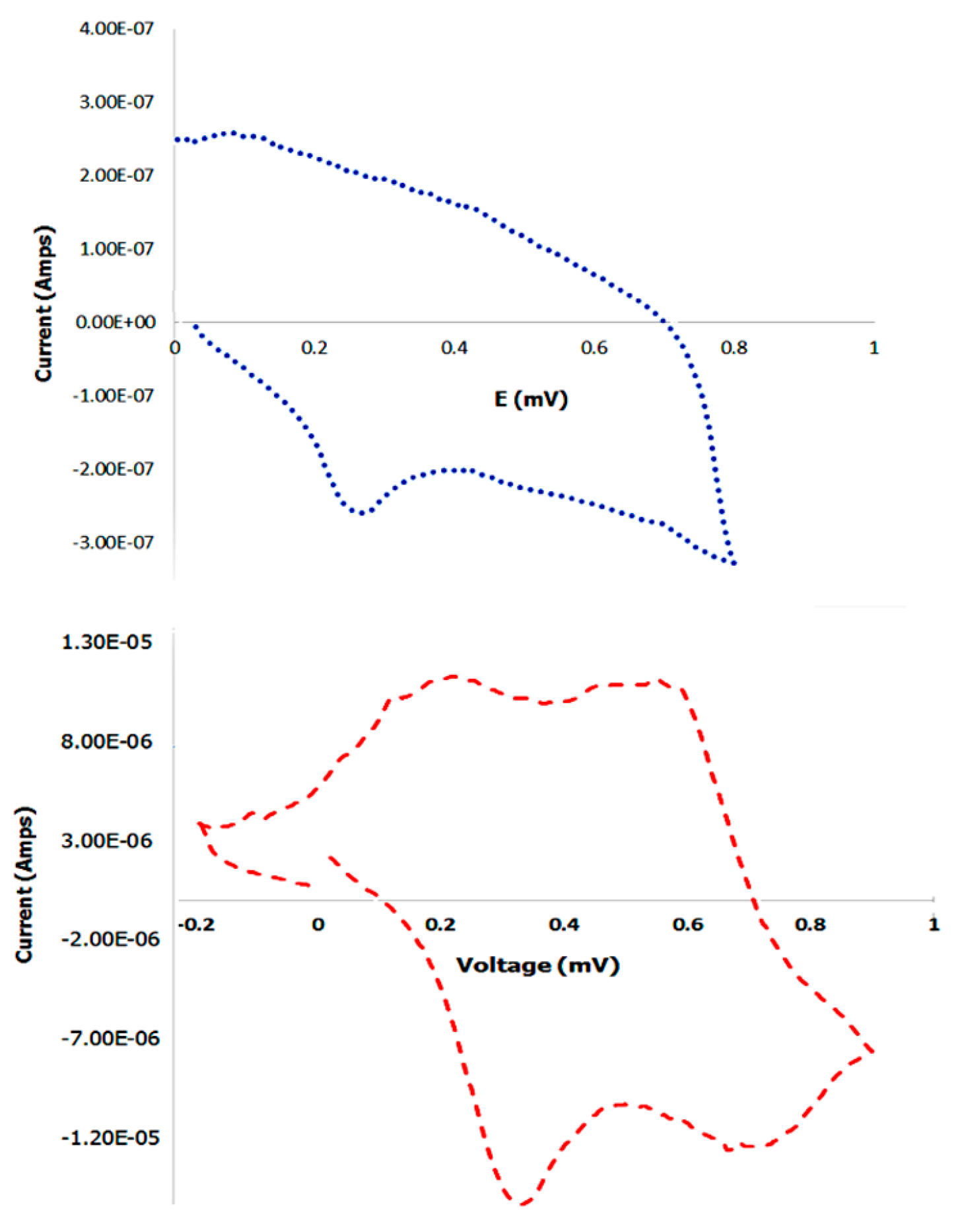
3.9. Cyclic Voltammetry
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Saaty, T. Neurons the decision makers, Part I: The firing function of a single neuron. Neural Netw. 2017, 86, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.; Cayre, M.; Panatier, A.; Avignone, E. Neuron-glial cell cooperation. In Cellular and Molecular Neurophysiology, 4th ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 25–37. ISBN 9780123970329. [Google Scholar]
- Katsu-Jiménez, Y.; Alves, R.M.P.; Giménez-Cassina, A. Food for thought: Impact of metabolism on neuronal excitability. Exp. Cell Res. 2017, 36, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Jayaram, S.; Madugundu, A.K.; Chavan, S.; Advani, J.; Pandey, A.; Thongboonkerd, V.; Sirdeshmukh, R. Chromosome-centric human proteome project: Deciphering proteins associated with glioma and neurodegenerative disorders on chromosome 12. J. Proteome Res. 2014, 13, 3178–3190. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Bellamkonda, R.V. Tissue-Engineered scaffolds are Effective alternatives to autografts for bridging peripheral nerve gaps. Tissue Eng. 2003, 9, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Otto, R.; Penzis, R.; Gaube, F.; Adolph, O.; Föhr, K.J.; Warncke, P.; Robaa, D.; Appenroth, D.; Fleck, C.; Enzensperger, C.; et al. Evaluation of homobivalent carbolines as designed multiple ligands for the treatment of neurodegenerative disorders. J. Med. Chem. 2015, 58, 6710–6715. [Google Scholar] [CrossRef] [PubMed]
- Carriel, V.; Alaminos, M.; Garzón, I.; Campos, A.; Cornelissen, M. Tissue engineering of the peripheral nervous system. Expert Rev. Neurother. 2014, 14, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Williams, D. Benefit and risk in tissue engineering. Mater. Today 2004, 7, 24–29. [Google Scholar] [CrossRef]
- Evans, N.D.; Gentleman, E.; Polak, J.M. Scaffolds for stem cells. Mater. Today 2006, 9, 26–33. [Google Scholar] [CrossRef]
- Gu, X.; Ding, F.; Williams, D.F. Neural tissue engineering options for peripheral nerve regeneration. Biomaterials 2014, 35, 6143–6156. [Google Scholar] [CrossRef] [PubMed]
- Koss, K.M.; Unsworth, L.D. Neural tissue engineering: Bioresponsive nanoscaffolds using engineered self-assembling peptides. Acta Biomater. 2016, 44, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Skop, N.B.; Calderon, F.; Cho, C.H.; Gandhi, C.D.; Levison, S.W. Improvements in biomaterial matrices for neural precursor cell transplantation. Mol. Cell. Ther. 2014, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Sato, Y.; Haniu, H.; Nomura, H.; Kobayashi, S.; Takanashi, S.; Okamoto, M.; Takizawa, T.; Aoki, K.; Usui, Y.; et al. A three-dimensional block structure consisting exclusively of carbon nanotubes serving as bone regeneration scaffold and as bone defect filler. PLoS ONE 2017, 12, e0172601. [Google Scholar] [CrossRef] [PubMed]
- Perkins, B.L.; Naderi, N. Carbon nanostructures in bone tissue engineering. Open Orthop. J. 2016, 10, 877–899. [Google Scholar] [CrossRef] [PubMed]
- Palejwala, A.H.; Fridley, J.S.; Mata, J.A.; Samuel, E.L.G.; Luerssen, T.G.; Perlaky, L.; Kent, T.A.; Tour, J.M.; Jea, A. Biocompatibility of reduced graphene oxide nanoscaffolds following acute spinal cord injury in rats. Surg. Neurol. Int. 2016, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Kohane, D.S.; Langer, R. Polymeric biomaterials in tissue engineering. Pediatr. Res. 2008, 63, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ushida, T.; Tateishi, T. Scaffold design for tissue engineering. Macromol. Biosci. 2002, 2, 67–77. [Google Scholar] [CrossRef]
- Fukunishi, T.; Shoji, T.; Shinoka, T. Nanofiber composites in vascular tissue engineering. In Nanofiber Composites for Biomedical Applications; Woodhead Publishing: Cambridge, UK, 2017; pp. 455–481. ISBN 9780081001738. [Google Scholar]
- Alvarez-Perez, M.A.; Guarino, V.; Cirillo, V.; Ambrosio, L. Influence of gelatin cues in PCL electrospun membranes on nerve outgrowth. Biomacromolecules 2010, 11, 2238–2246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, R.; Wang, Z.; Dong, M.; Cui, B.; Chen, M. Visible-Light neural stimulation on graphitic-carbon nitride/graphene photocatalytic fibers. ACS Appl. Mater. Interfaces 2017, 9, 34736–34743. [Google Scholar] [CrossRef] [PubMed]
- Hemshekhar, M.; Thushara, R.M.; Chandranayaka, S.; Sherman, L.S.; Kemparaju, K.; Girish, K.S. Emerging roles of hyaluronic acid bioscaffolds in tissue engineering and regenerative medicine. Int. J. Biol. Macromol. 2016, 86, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, K.C.; Conovaloff, A.W.; Panitch, A. Development of affinity-based delivery of NGF from a chondroitin sulfate biomaterial. Biomatter 2011, 1, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Weyers, A.; Linhardt, R.J. Neoproteoglycans in tissue engineering. FEBS J. 2013, 280, 2511–2522. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Yuan, Y.; Lyu, Z.; Wang, M.; Liu, Q.; Wang, H.; Yuan, L.; Chen, H. Deciphering the role of sulfonated unit in heparin-mimicking polymer to promote neural differentiation of embryonic stem cells. ACS Appl. Mater. Interfaces 2017, 9, 28209–28221. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Thouas, G.A.; Bernard, C.C.; Nisbet, D.R.; Finkelstein, D.I.; Li, D.; Forsythe, J.S. Method to impart electro- and biofunctionality to neural scaffolds using graphene-polyelectrolyte multilayers. ACS Appl. Mater. Interfaces 2012, 4, 4524–4531. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Gilbert, R.J.; He, W. Simple agarose-chitosan gel composite system for enhanced neuronal growth in three dimensions. Biomacromolecules 2009, 10, 2954–2959. [Google Scholar] [CrossRef] [PubMed]
- Entekhabi, E.; Haghbin Nazarpak, M.; Moztarzadeh, F.; Sadeghi, A. Design and manufacture of neural tissue engineering scaffolds using hyaluronic acid and polycaprolactone nanofibers with controlled porosity. Mater. Sci. Eng. C 2016, 69, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Rad-Malekshahi, M.; Lempsink, L.; Amidi, M.; Hennink, W.E.; Mastrobattista, E. Biomedical Applications of Self-Assembling Peptides. Bioconjug. Chem. 2016, 27, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Mazza, M.; Hadjidemetriou, M.; De Lázaro, I.; Bussy, C.; Kostarelos, K. Peptide nanofiber complexes with siRNA for deep brain gene silencing by stereotactic neurosurgery. ACS Nano 2015, 9, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Stephanopoulos, N.; Freeman, R.; North, H.A.; Sur, S.; Jeong, S.J.; Tantakitti, F.; Kessler, J.A.; Stupp, S.I. Bioactive DNA-peptide nanotubes enhance the differentiation of neural stem cells into neurons. Nano Lett. 2015, 15, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Behnke, R.G.; Liang, Y.-X.; You, S.-W.; Tay, D.K.C.; Zhang, S.; So, K.-F.; Schneider, G.E. Nano neuro knitting: peptide nanofiber scaffold for brain repair and axon regeneration with functional return of vision. Proc. Natl. Acad. Sci. USA 2006, 103, 5054–5059. [Google Scholar] [CrossRef] [PubMed]
- Sur, S.; Pashuck, E.T.; Guler, M.O.; Ito, M.; Stupp, S.I.; Launey, T. A hybrid nanofiber matrix to control the survival and maturation of brain neurons. Biomaterials 2012, 33, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Hutton, J.C.; Schofield, P.J.; Williams, J.F.; Hollows, F.C. The failure of aldose reductase inhibitor 3,3′-tetramethylene glutaric acid to inhibit in vivo sorbitol accumulation in lens and retina in diabetes. Biochem. Pharmacol. 1974, 23, 2991–2998. [Google Scholar] [CrossRef]
- Rosenberyg, D.; Artoul, S.; Segal, A.C.; Kolodney, G.; Radzishevsky, I.; Dikpoltsev, E.; Foltyn, V.N.; Inoue, R.; Mori, H.; Billard, J.-M.; et al. Neuronal D-Serine and Glycine Release Via the Asc-1 Transporter Regulates NMDA Receptor-Dependent Synaptic Activity. J. Neurosci. 2013, 33, 3533–3544. [Google Scholar] [CrossRef] [PubMed]
- Arulmoli, J.; Pathak, M.M.; McDonnell, L.P.; Nourse, J.L.; Tombola, F.; Earthman, J.C.; Flanagan, L.A. Static stretch affects neural stem cell differentiation in an extracellular matrix-dependent manner. Sci. Rep. 2015, 5, 8499. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.; Yeon Kim, J.; Lee, E.; Hong, N.; Sun, W.; Nam, Y. Effects of ECM protein micropatterns on the migration and differentiation of adult neural stem cells. Sci. Rep. 2015, 5, 13043. [Google Scholar] [CrossRef] [PubMed]
- Junka, R.; Valmikinathan, C.M.; Kalyon, D.M.; Yu, X. Laminin functionalized biomimetic nanofibers for nerve tissue engineering. J. Biomater. Tissue Eng. 2013, 3, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Tate, C.C.; Shear, D.A.; Tate, M.C.; Archer, D.R.; Stein, D.G.; LaPlaca, M.C. Laminin and fibronectin scaffolds enhance neural stem cell transplantation into the injured brain. J. Tissue Eng. Regen. Med. 2009, 3, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Baloh, R.H.; Tansey, M.G.; Lampe, P.A.; Fahrner, T.J.; Enomoto, H.; Simburger, K.S.; Leitner, M.L.; Araki, T.; Johnson, E.M.; Milbrandt, J. Artemin, a novel member of the GDNF ligand family, supports peripheral and central neurons and signals through the GFRα3-RET receptor complex. Neuron 1998, 21, 1291–1302. [Google Scholar] [CrossRef]
- Detloff, M.R.; Smith, E.J.; Quiros Molina, D.; Ganzer, P.D.; Houlé, J.D. Acute exercise prevents the development of neuropathic pain and the sprouting of non-peptidergic (GDNF- and artemin-responsive) c-fibers after spinal cord injury. Exp. Neurol. 2014, 255, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.J.; Watson, J.J.; Shoemark, D.K.; Barua, N.U.; Patel, N.K. GDNF, NGF and BDNF as therapeutic options for neurodegeneration. Pharmacol. Ther. 2013, 138, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Timpl, R.; Wiedmann, H.; van Delden, V.; Furthmayr, H.; Kűhn, K. A network model for the organization of type IV collagen molecules in basement membranes. Eur. J. Biochem. 1981, 120, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Boeva, Z.A.; Sergeyev, V.G. Polyaniline: Synthesis, properties and application. Polym. Sci. Ser. C 2014, 56, 144–153. [Google Scholar] [CrossRef]
- Magnuson, M.; Guo, J.-H.; Butorin, S.M.; Agui, A.; Sathe, C.; Nordgren, J. The electronic structure of polyaniline and doped phases studied by soft X-ray absorption and emission spectroscopies. J. Chem. Phys. 1999, 111, 4756–4763. [Google Scholar] [CrossRef]
- Li, M.; Guo, Y.; Wei, Y.; MacDiarmid, A.G.; Lelkes, P.I. Electrospinning polyaniline-contained gelatin nanofibers for tissue engineering applications. Biomaterials 2006, 27, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Shi, J.; Zhu, M.; Chen, Y.; Chen, F. The three-stage in vitro degradation behavior of mesoporous silica in simulated body fluid. Microporous Mesoporous Mater. 2010, 131, 314–320. [Google Scholar] [CrossRef]
- Sun, L.; Zheng, C.; Webster, T.J. Self-assembled peptide nanomaterials for biomedical applications: Promises and pitfalls. Int. J. Nanomed. 2017, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Borland, S.; Richardson, S.M.; Merry, C.L.R.; Saiani, A.; Gough, J.E. Self-assembling peptide hydrogel for intervertebral disc tissue engineering. Acta Biomater. 2016, 46, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Kogiso, M.; Ohnishi, S.; Yase, K.; Masuda, M.; Shimizu, T. Dicarboxylic oligopeptide bolaamphiphiles: Proton-triggered self-assembly of microtubes with loose solid surfaces. Langmuir 1998, 14, 4978–4986. [Google Scholar] [CrossRef]
- Menzenski, M.Z.; Banerjee, I.A. Self-assembly of supramolecular nanostructures from phenylalanine derived bolaamphiphiles. New J. Chem. 2007, 31, 1674. [Google Scholar] [CrossRef]
- Cote, Y.; Fu, I.W.; Dobson, E.T.; Goldberger, J.E.; Nguyen, H.D.; Shen, J.K. Mechanism of the pH-controlled self-assembly of nanofibers from peptide amphiphiles. J. Phys. Chem. C 2014, 118, 16272–16278. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Gologan, B. Crystalline glycylglycine bolaamphiphile tubules and their pH-sensitive structural transformation. J. Phys. Chem. B 2000, 104, 3383–3386. [Google Scholar] [CrossRef]
- Loo, Y.; Zhang, S.; Hauser, C.A.E. From short peptides to nanofibers to macromolecular assemblies in biomedicine. Biotechnol. Adv. 2012, 30, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Cuvier, A.-S.; Berton, J.; Stevens, C.V.; Fadda, G.C.; Babonneau, F.; Van Bogaert, I.N.A.; Soetaert, W.; Pehau-Arnaudet, G.; Baccile, N. pH-triggered formation of nanoribbons from yeast-derived glycolipid biosurfactants. Soft Matter 2014, 10, 3950–3959. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, P.R.; Lustig, A.; Steinmetz, M.O.; Kammerer, R.A. Laminin chain assembly is regulated by specific coiled-coil interactions. J. Struct. Biol. 2010, 170, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.-A.; Carafoli, F.; Hohenester, E. Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region. EMBO Rep. 2011, 12, 276–282. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, J.; Amons, R.; Möller, W. The primary structure of artemin from Artemia cysts. Eur. J. Biochem. 1990, 193, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Hassani, L.; Sajedi, R.H. Effect of artemin on structural transition of lactoglobulin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 105, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Agarwal, R.; Ko, K.R.; Ruthven, M.; Sarhan, H.T.; Frampton, J.P. Templated assembly of collagen fibers directs cell growth in 2D and 3D. Sci. Rep. 2017, 7, 9628. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Wan, M. Self-assembled polyaniline nanostructures with photoisomerization function. Chem. Mater. 2002, 14, 3486–3492. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. (Shanghai) 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Almlén, A.; Vandenbussche, G.; Linderholm, B.; Haegerstrand-Björkman, M.; Johansson, J.; Curstedt, T. Alterations of the C-terminal end do not affect in vitro or in vivo activity of surfactant protein C analogs. Biochim. Biophys. Acta. Biomembr. 2012, 1818, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Langham, A.A.; Waring, A.J.; Kaznessis, Y.N. Comparison of interactions between beta-hairpin decapeptides and SDS/DPC micelles from experimental and simulation data. BMC Biochem. 2007, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.R.; Garrell, R.L. g-Factor analysis of protein secondary structure in solutions and thin films. Faraday Discuss. 2004, 126, 209. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Wang, Y.; Guo, Z.; Meng, H.; Huang, J.; Zhang, L.; Zhao, B.; Zhao, Q.; Zheng, Y.; Peng, J. Cauda equina-derived extracellular matrix for fabrication of nanostructured hybrid scaffolds applied to neural tissue engineering. Tissue Eng. Part A 2015, 21, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Vass, E.; Hollósi, M.; Besson, F.; Buchet, R. Vibrational spectroscopic detection of beta- and gamma-turns in synthetic and natural peptides and proteins. Chem. Rev. 2003, 103, 1917–1954. [Google Scholar] [CrossRef] [PubMed]
- Pud, A.A.; Nikolayeva, O.A.; Vretik, L.O.; Noskov, Y.V.; Ogurtsov, N.A.; Kruglyak, O.S.; Fedorenko, E.A. New nanocomposites of polystyrene with polyaniline doped with lauryl sulfuric acid. Nanoscale Res. Lett. 2017, 12, 493. [Google Scholar] [CrossRef] [PubMed]
- Arruebo, M.; Valladares, M.; Gonzalez-Fernandez, A. Antibody conjugated nanoparticles for biomedical applications. J. Nanomaterials 2009, 2009, 37. [Google Scholar] [CrossRef]
- Gröschel, A.H.; Müller, A.H.E. Self-assembly concepts for multicompartment nanostructures. Nanoscale 2015, 7, 11841–11876. [Google Scholar] [CrossRef] [PubMed]
- Heim, A.J.; Matthews, W.G.; Koob, T.J. Determination of the elastic modulus of native collagen fibrils via radial indentation. Appl. Phys. Lett. 2006, 89. [Google Scholar] [CrossRef]
- Muiznieks, L.D.; Keeley, F.W. Molecular assembly and mechanical properties of the extracellular matrix: A fibrous protein perspective. Biochim. Biophys. Acta 2012, 1832, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Li, X.; Wang, X.; Zhang, J.; Yu, D.; Qiu, B. Fabrication of vertical array CNTs/polyaniline composite membranes by microwave-assisted in situ polymerization. Nanoscale Res. Lett. 2015, 10, 493. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jang, G.W.; Hsueh, K.F.; Scherr, E.M.; MacDiarmid, A.G.; Epstein, A.J. Thermal transitions and mechanical properties of films of chemically prepared polyaniline. Polymer (Guildf) 1992, 33, 314–322. [Google Scholar] [CrossRef]
- Xu, C.; Yang, F.; Wang, S.; Ramakrishna, S. In vitro study of human vascular endothelial cell function on materials with various surface roughness. J. Biomed. Mater. Res. A 2004, 71, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-Q.; Kankala, R.K.; Chen, A.-Z.; Yang, D.-Z.; Cheng, X.-X.; Jiang, N.-N.; Zhu, K.; Wang, S.-B. Investigation of silk fibroin nanoparticle-decorated poly(l-lactic acid) composite scaffolds for osteoblast growth and differentiation. Int. J. Nanomed. 2017, 12, 1877–1890. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.T.; West, J.L. Photopolymerizable hydrogels for tissue engineering applications. Biomaterials 2002, 23, 4307–4314. [Google Scholar] [CrossRef]
- Sheikh, Z.; Najeeb, S.; Khurshid, Z.; Verma, V.; Rashid, H.; Glogauer, M. Biodegradable materials for bone repair and tissue engineering applications. Materials (Basel) 2015, 8, 5744–5794. [Google Scholar] [CrossRef] [PubMed]
- Serbo, J.V.; Gerecht, S. Vascular tissue engineering: biodegradable scaffold platforms to promote angiogenesis. Stem Cell Res. Ther. 2013, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- Ikejima, T.; Inoue, Y. Crystallization behavior and environmental biodegradability of the blend films of poly(3-hydroxybutyric acid) with chitin and chitosan. Carbohydr. Polym. 2000, 41, 351–356. [Google Scholar] [CrossRef]
- Bober, P.; Humpolíček, P.; Pacherník, J.; Stejskal, J.; Lindfors, T. Conducting polyaniline based cell culture substrate for embryonic stem cells and embryoid bodies. RSC Adv. 2015, 5, 50328–50335. [Google Scholar] [CrossRef]
- Neal, R.A.; Lenz, S.M.; Wang, T.; Abebayehu, D.; Brooks, B.P.C.; Ogle, R.C.; Botchwey, E.A. Laminin- and basement membranepolycaprolactone blend nanofibers as a scaffold for regenerative medicine. Nanomater. Environ. 2014, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.E.; Gibson, M.E.; Arnold, H.M.; Pepinsky, B.; Frank, E. Artemin promotes functional long-distance axonal regeneration to the brainstem after dorsal root crush. Proc. Natl. Acad. Sci. USA 2015, 112, 6170–6175. [Google Scholar] [CrossRef] [PubMed]
- Önger, M.E.; Delibaş, B.; Türkmen, A.P.; Erener, E.; Altunkaynak, B.Z.; Kaplan, S. The role of growth factors in nerve regeneration. Drug Discov. Ther. 2016, 3, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.J.; Kim, S.H.; Yang, D.H.; Kwon, H.; Shin, J.S. Amperometric glucose biosensor by means of electrostatic layer-by-layer adsorption onto polyaniline-coated polyester films. J. Ind. Eng. Chem. 2010, 16, 380–384. [Google Scholar] [CrossRef]
- Ku, S.H.; Lee, S.H.; Park, C.B. Synergic effects of nanofiber alignment and electroactivity on myoblast differentiation. Biomaterials 2012, 33, 6098–6104. [Google Scholar] [CrossRef] [PubMed]
- Gajendran, P.; Saraswathi, R. Polyaniline-carbon nanotube composites. Pure Appl. Chem. 2008, 80, 2377–2395. [Google Scholar] [CrossRef]

| Construct | Young’s Modulus (GPa) |
|---|---|
| Self-Assembled nanoribbons | 0.757 |
| Nanoribbons bound to Laminin | 1.805 |
| Nanoribbons bound to Laminin-Artemin-Collagen | 2.985 |
| Nanoribbons bound to Laminin-Artemin-Collagen-PANI | 5.522 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, A.M.; Pajovich, H.T.; Banerjee, I.A. Development of Self-Assembled Nanoribbon Bound Peptide-Polyaniline Composite Scaffolds and Their Interactions with Neural Cortical Cells. Bioengineering 2018, 5, 6. https://doi.org/10.3390/bioengineering5010006
Smith AM, Pajovich HT, Banerjee IA. Development of Self-Assembled Nanoribbon Bound Peptide-Polyaniline Composite Scaffolds and Their Interactions with Neural Cortical Cells. Bioengineering. 2018; 5(1):6. https://doi.org/10.3390/bioengineering5010006
Chicago/Turabian StyleSmith, Andrew M., Harrison T. Pajovich, and Ipsita A. Banerjee. 2018. "Development of Self-Assembled Nanoribbon Bound Peptide-Polyaniline Composite Scaffolds and Their Interactions with Neural Cortical Cells" Bioengineering 5, no. 1: 6. https://doi.org/10.3390/bioengineering5010006





