1. Introduction
De novo methods for drug discovery can be incredibly expensive and time consuming. A target molecule, such as a gene that is causally linked to a disease, is first identified to initiate the drug discovery process. Scientists then systematically screen for small molecules that modulate the target protein-product and carry out a series of optimization steps to improve the efficacy of the lead drug. Next, periods of animal studies are followed by clinical trials before a drug can be approved for the market by the USA Food and Drug Administration (FDA) (or its counterpart in other countries). Usually, this procedure costs four to 12 billion dollars and takes an average of 12 to 17 years to complete [
1]. Unfortunately, in some cases, this may not result in any successful drug that is safe for human use. For example, if a promising drug starts to reveal severe side effects during clinical trials, the drug is discarded, and the resources invested would thus be wasted. Due to these limitations, only 50 new drugs could be approved by the U.S. FDA during a period of ten years from 1999 to 2008 [
2].
In order to make the current drug discovery practices more efficient or productive, a new approach based on repurposing the existing FDA-approved drugs for new diseases has recently gained traction [
1]. Drug repurposing, which is typically based on systems biology and data analytics, could drastically reduce both the cost and time associated with finding suitable drugs for diseases. There are several drugs that were successfully repurposed to treat new diseases [
3], of which some well-known examples include: anti-angina drug sildenafil citrate repurposed to treat erectile dysfunction [
4], the diabetes medicine metformin that has potential use for cancer treatment [
5], HIV drug nucleoside reverse transcriptase inhibitors that could treat “dry” age-related macular degeneration [
6] and imipramine, a tricyclic antidepressant, with potential to treat small-cell lung carcinoma [
7].
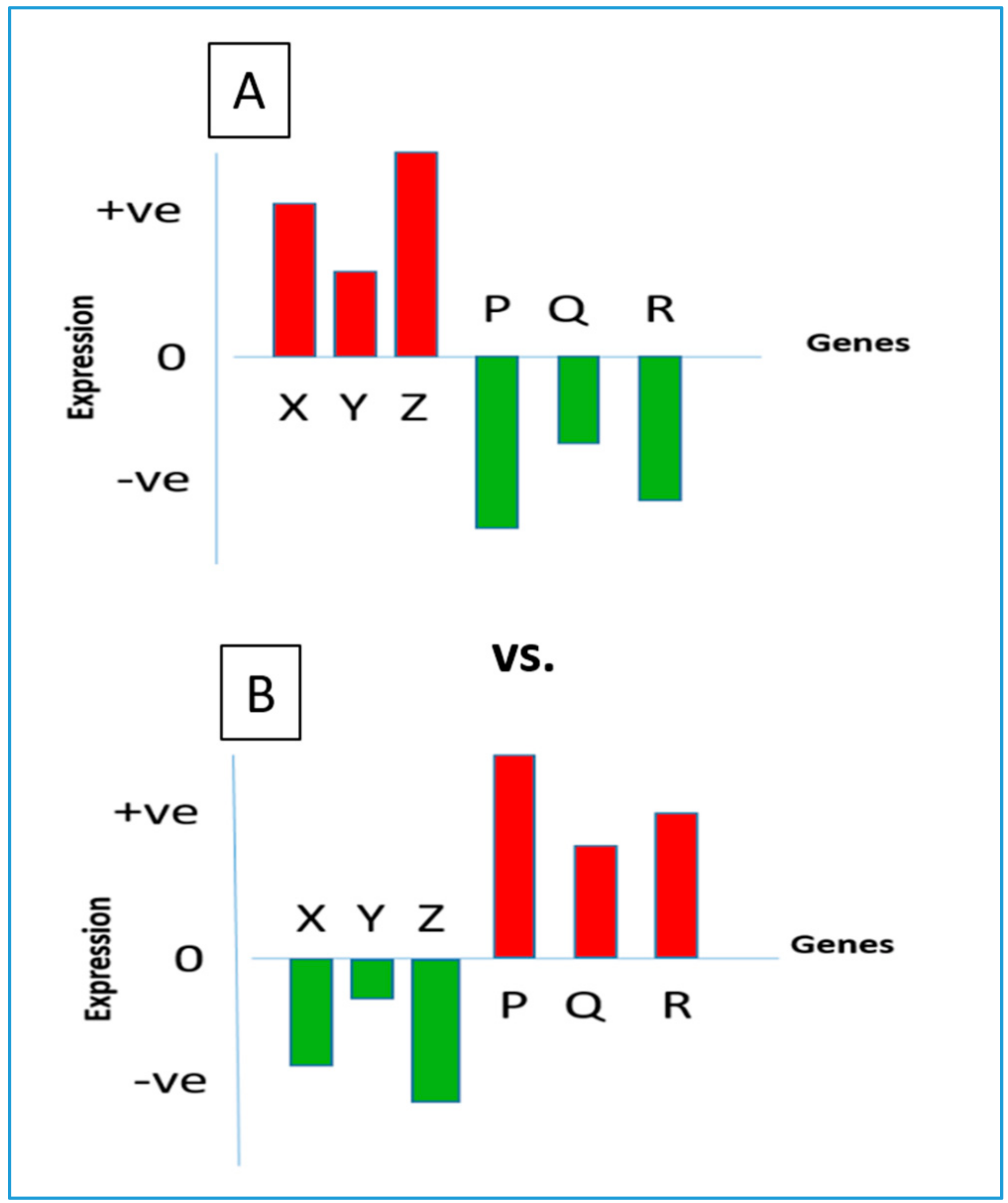
A common method of identifying repurposed drugs is based on the following principle. Human cells maintain homeostasis through the regulated expression of their transcriptome. A perturbation of cellular gene expression often leads to the manifestation of a disease state; hence, each disease normally reveals a signature gene expression profile [
8]. When a suitable drug is administered to treat a disease, it tends to correct the aberration by bringing the gene expression pattern back to its normal state. Thus, a negative correlation is generally revealed between the gene expression signatures of the drug and the disease [
8,
9]. A schematic diagram representing opposing gene expression patterns under a disease state and under its drug treatment is shown in
Figure 1.
Advancements in high-throughput technologies, such as microarrays and RNA-sequencing methods, allow scientists to measure gene expression patterns under different experimental conditions. Scientists routinely deposit gene expression data derived from such experiments into online repositories, often times making the data freely available [
10].
Published studies have shown that the statistical analysis of gene expression data can be utilized to identify repurposable drugs [
8]. However, running these sophisticated correlation statistics on gene expression datasets requires proficiency in the fields of computer science, bioinformatics and statistics, along with access to high powered computers. The advent of bioinformatics tools that mine raw gene expression data from the repository and run sophisticated statistical algorithms plays an important role in making pre-processed transcriptomics data available to the public.
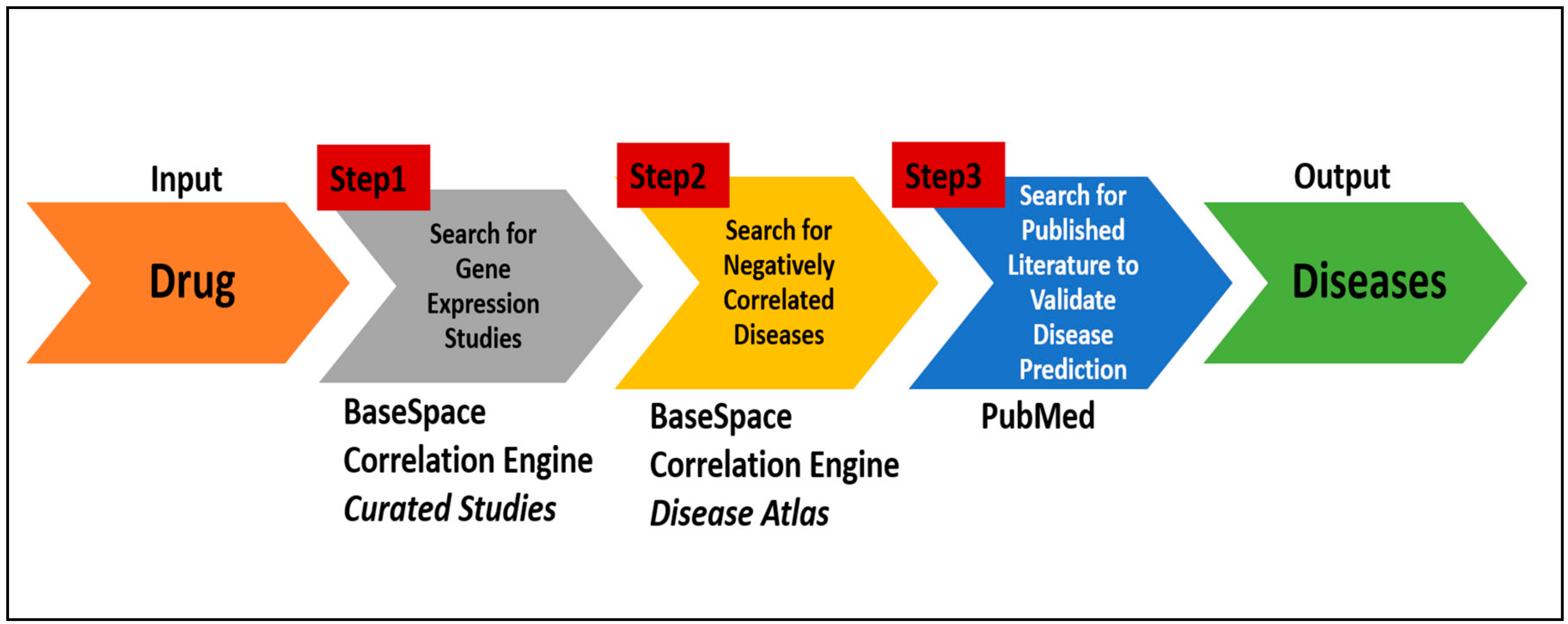
We have developed a protocol by sequentially connecting disparate existing online bioinformatics tools, databases and literature search engines to identify repurposable drugs. This protocol, which can be used by any motivated individual, takes an FDA approved drug as the input and then searches for diseases that show negatively correlated gene expression in relation to that drug. The efficacy of this protocol was assessed by comparing the protocol prediction with the results of two recently-published articles of repurposed drugs: zidovudine [
6] and imipramine [
7].
2. Methods
2.1. BaseSpace Correlation Engine
The BaseSpace Correlation Engine utilizes proprietary statistical algorithms to convert raw experimental data into a list of genes that are differentially expressed in certain conditions along with their corresponding fold change and p-value calculations. The fold change value indicates how a given gene is differentially expressed in a test condition compared to the control condition of an experiment. Examples are experiments with drug treatment vs. non-treatment or disease state vs. normal state. Rank-based enrichment statistics are then used to compute the pairwise correlation scores between all gene expression signatures present in the database. The most correlated gene expression study present for each query was assigned a numerical score of 100, and scores for the rest of the results were normalized to the top-ranked study. This resource enables users to find gene expression experiments with given drug-treated versus untreated conditions. Hence, diseases showing strong negative correlation with a given drug can be identified to predict potentially repurposable drugs.
2.2. PubMed
An online database (
https://www.ncbi.nlm.nih.gov/pubmed) is comprised of over 26 million citations published in 2600 life sciences journals. Many of these citations provide links to the abstract and full text of the article. This database is freely accessible and maintained by the National Library of Medicine, a division of the U.S. National Institute of Health.
2.3. In Silico Protocol to Identify Repurposed Drugs
A protocol encompassing the systematic search of the online databases mentioned above enables users to quickly and efficiently identify potential target diseases for a drug (
Figure 2). The steps are as follows:
a. Identify gene expression studies associated with the drug:
Enter a drug name in the search box of the BaseSpace Correlation Engine and then click on the icon named “Curated Studies” displayed at the top of the web page. Once the search result is returned, click on the “Filter By” option and select “Data Types” available at the top of the page and select “RNA Expression.”
Browse the search results returned that will display numerous independent gene expression studies. Identify the studies that compare the gene expressed data for drug treatment vs. control (untreated). Select the appropriate study by clicking on the hyperlink of that study. Each study may have multiple experiments measured under different conditions such as a different dosage of the drug or different treatment time points. Clicking on the study will bring up a page with a detailed description of the study, as well as links to the gene expression data associated with each experiment. When the experiment was selected by clicking the hyperlinked title and a table consisting of the differential gene expression, data will appear on the screen.
b. Search for diseases with negative correlations:
Select the icon “Disease Atlas” available at the top of the page. A web page displaying a table of various disease names with their corresponding correlation scores will appear. Sort through the table by selecting “Rank” from the drop-down menu available under the “View By” option displayed at the top left corner of the page. Re-sort the results by selecting the “−ve Correlation” option present in the drop-down menu under the “Correlation with Query” column heading at the top of the right-most column. Finally, select the diseases that have the largest number of studies. The queried drug would have the potential to treat the selected diseases.
c. Search literature database to validate disease predictions:
Go to PubMed, and enter “the drug name AND the predicted disease name” in the search box and look for citations.
3. Results
This protocol was applied to two previously published repurposed drugs, and the protocol predictions were compared with the reported findings.
3.1. Case Study 1: Zidovudine
Through experiments performed in mouse models, Fowler et al. reported that the drug zidovudine, a Nucleoside Reverse Transcriptase Inhibitor (NRTI) usually used to treat HIV patients, showed strong potential to be used against the untreatable dry form of Age-related Macular Degeneration (AMD) [
6].
3.1.1. Study Selection from the BaseSpace Correlation Engine
A search for curated studies using the query zidovudine in the BaseSpace Correlation Engine retrieved 10 RNA-expression studies: six studies based on rat experiments and four from experiments done on human data. A study titled “Drug Matrix In Vitro Toxicogenomic Study—Rat Hepatocytes [Affymetrix]” [
12,
13] was selected for further analysis. Although drug-treatment studies performed on normal human cell lines would be preferred for repurposed disease prediction, for this query, all human studies were only carried out for cancer cell lines or virus-infected cells. Hence, they were not suitable for study selection. On the other hand, the studies based on rat experiments were performed under normal conditions. Since the liver is a principal site of drug metabolism, a study measuring the gene expression patterns of rat hepatocyte cells was selected.
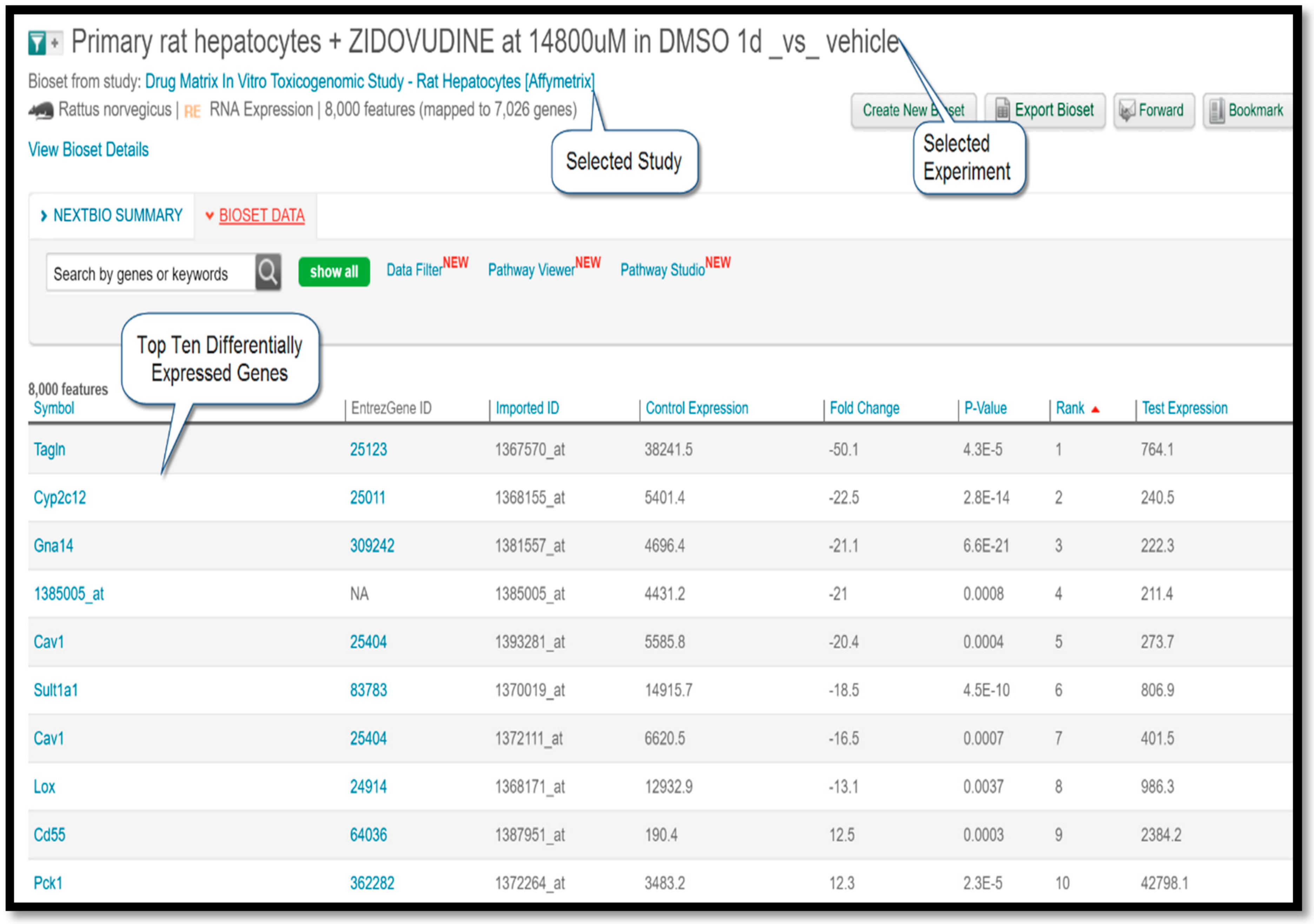
In this study, rat hepatocyte cells isolated from male Sprague-Dawley rats were co-cultured in vitro with varying doses of the drug during different time points. Microarray experiments were then performed on an Affymetrix platform to measure and compare the gene expression patterns of treatment versus control conditions. Typically, a gene expression study is comprised of multiple experiments. Among the three experiments listed under this study, the experiment titled “Primary rat hepatocytes + ZIDOVUDINE at 14,800 µM in DMSO 1D_vs_vehicle” covered the largest number of genes (8000). Therefore, the differentially-expressed gene (DEG) profile of the said experiment was used as a query to seek out Negatively-Correlated (NC) diseases for zidovudine.
Figure 3 shows a screenshot taken from the BaseSpace Correlation Engine result page displaying the top ten differentially-expressed genes.
3.1.2. Negatively-Correlated Diseases
The BaseSpace Correlation Engine ranks the negatively-correlated diseases based on its assigned correlation score. The most correlated gene expression study, with the lowest
p-value, present for each query is assigned a numerical score of 100 and scores for the rest of the results were normalized to the top-ranked study. However, for this protocol, the ranking was manually changed to reflect the number of supporting, independent studies. This step was taken to bolster the prediction efficiency with the notion that if more independent experiments found a negative correlation between the drug and the disease, then its ranking should be stronger (while still maintaining an acceptable score of at least 50/100 to keep correlation as a factor).
Table 1 shows the top 10 negatively-correlated diseases for HIV-drug zidovudine ordered by the number of supported gene expression studies.
Among the diseases listed in the table, human immunodeficiency virus infection, the intended target of zidovudine, is present (No. 5). The gene expression profiles derived from twenty-one studies revealed a strong statistically-significant negative correlation (score: 62) with the expression pattern of zidovudine.
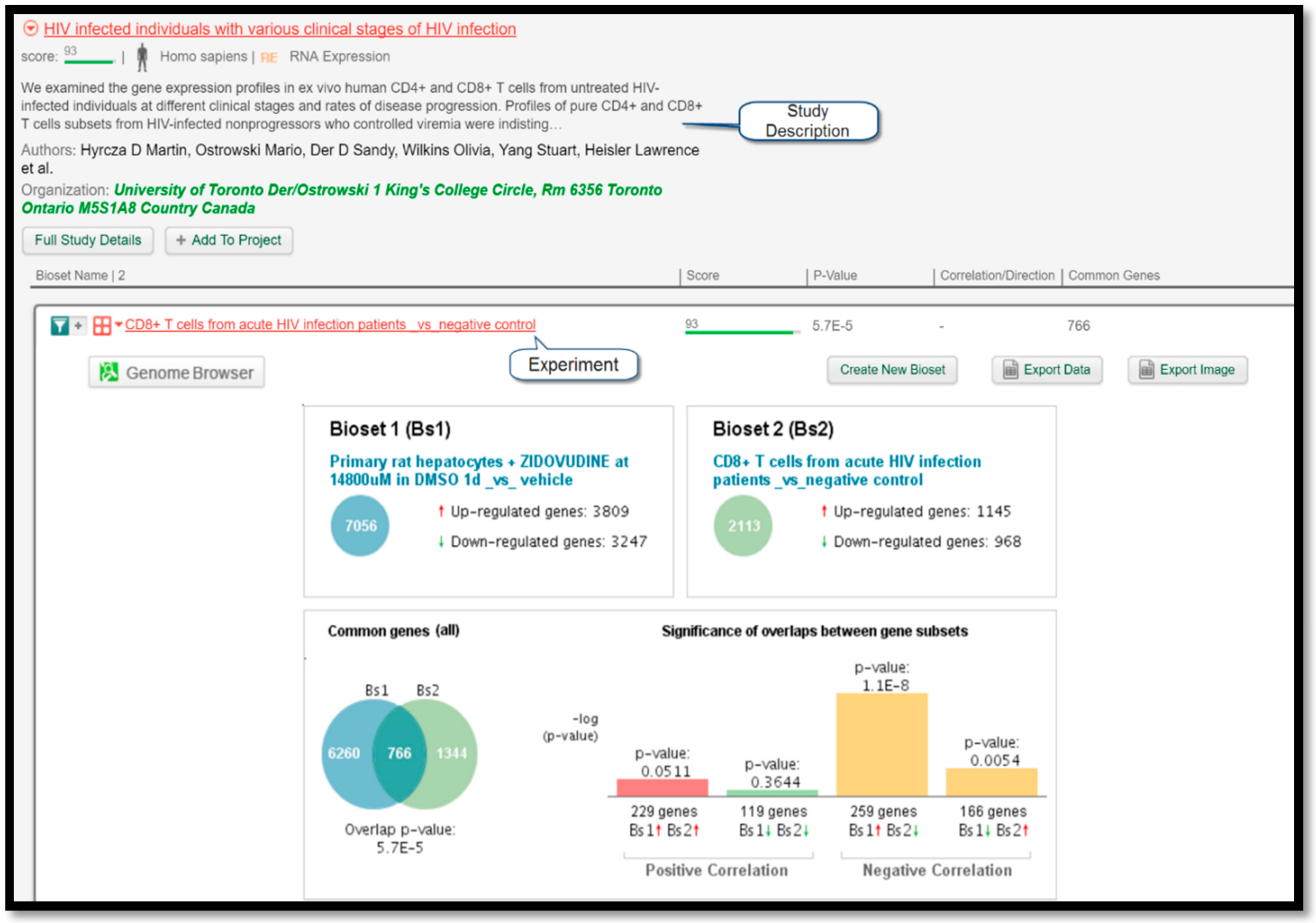
Figure 4 displays a comparison of gene expressions between the query experiment and the experiment “CD8+ T cells from chronic HIV infection patients _vs_ negative control” listed under the study “HIV-infected individuals with various clinical stages of HIV infection” performed by Hyrcza et al. [
16]. Since the protocol correctly predicted that zidovudine could treat HIV, its intended target, this finding serves as a valuable result to prove the efficacy of this protocol.
For Age-related Macular Degeneration (AMD), the published finding [
6] that we sought falls under the broader term “retinal disorder,” which is present in the list of the NC diseases (No. 9). The DEG profile of the experiment “Macular retina—GA Age-related macular degeneration vs. normal tissue” as part of the study “Age-related macular degeneration subtype expression analysis” revealed a strong negative correlation with the gene expression profile of zidovudine [
17].
Apart from HIV, the intended target of zidovudine, all other NC diseases are novel findings, as they are not listed as a therapeutic target of zidovudine in well-known drug information resources, such as DrugBank [
18] and the National Library of Medicine Drug Information Portal [
19]. While the potential of this drug to treat AMD has been recently published, a literature search was conducted to collect evidence on whether the other NC diseases found have been connected to zidovudine. A literature search found Marcais et al. reported that the treatment of zidovudine is highly effective in the treatment of a leukemic subtype of adult t-cell lymphoma [
15]. Furthermore, Beck-Engeser et al. published a paper on the efficacy of zidovudine treatment against lupus erythematosus [
14]. However, no literature evidence was found for cardiovascular disease, neuropathy, mycobacteriosis, rheumatoid arthritis, myopathy or dermatitis.
3.2. Case Study 2: Imipramine
In 2013, Jahchan and colleagues reported that the Tricyclic Antidepressant (TCA) imipramine could be efficiently repurposed to treat small-cell lung carcinoma [
7]. In a protocol of their own, this study sought small molecules with the ability to treat the apparently recalcitrant form of lung cancer first by analyzing the disease-derived transcriptomic data and then by running experiments in an animal model. One of the small molecules identified was imipramine.
3.2.1. Study/Experiment Selection from BaseSpace Correlation Engine
A search for Imipramine (performed on 20 June 2016) in the BaseSpace Correlation Engine results in eight studies. The DEG profile derived from the experiment titled “Hepatocytes of female donors treated 24hr with 15uM imipramine _vs_ 0uM” done in human as a part of the study “Genomics Assisted Toxicity Evaluation system study—Human Hepatocytes” was selected for target disease prediction through this protocol [
20].
3.2.2. Negatively Correlated Diseases:
A list of NC diseases based on the DEG profile expressed by the selected query experiment is presented in
Table 2. The list shows a preponderance of cancer subtypes. Note that small-cell lung carcinoma, the reported therapeutic target for imipramine, is present in the NC disease list under lung cancer (No. 3). Hence, the findings reported by Jahchan et al. [
7] were successfully replicated by this in silico protocol. A literature search on imipramine retrieved previously published articles that studied the treatment of imipramine on two of the predicted target diseases: breast cancer [
21] and brain cancer [
22]. However, no supporting evidence from the literature was found for the treatment of liver cancer, kidney cancer, inflammatory bowel disease or Severe Acute Respiratory Syndrome (SARS) using imipramine. The prediction of SARS is notable because it revealed the highest correlation score of 100. However, the lack of reported evidence on the repositioning of imipramine against SARS further emphasizes the novelty of this prediction.
4. Conclusions
We have been able to verify the findings of previously reported results for two drugs: HIV-drug zidovudine and tricyclic antidepressant drug imipramine. Through this protocol, HIV-treating drug zidovudine was found to have the potential to be repurposed as a drug for dry age-related macular degeneration [
17] along with several other diseases, including lupus erythematosus, cardiovascular disease, lymphoid leukemia, neuropathy, mycobacteriosis, rheumatoid arthritis and dermatitis. Among these, four of the findings were supported by evidence from published literature. Imipramine could have the potential to treat small-cell lung carcinoma [
7]. Furthermore, we found that imipramine has the potential to treat several other diseases, including breast cancer, allergic disorder, liver cancer, brain cancer, inflammatory bowel disorder, cardiomyopathy, kidney cancer, nerve injury and Severe Acute Respiratory Syndrome (SARS). Five of the results found have been supported with published literature.
5. Limitations
This online repurposed drug prediction protocol depends on the availability of access to the BaseSpace Correlation Engine and its analysis of transcriptomics data. If the data for a given drug are not available, the protocol cannot be applied to that drug.
Furthermore, the DEG data gathered from experiments that compare the effect of the drug treatment on a normal (non-diseased) human cell line or tissue to the untreated or vehicle treated condition are considered to be ideal datasets for this method. However, for many drugs, such datasets are not available. Instead, the available DEG data come from experiments on human cancer cell lines or from experiments performed on animal models, such as mouse and rat. The protocol utilizing the non-ideal dataset may infer erroneous predictions and requiring additional analysis.
The BaseSpace Correlation Engine was freely available to the research community, and the authors received free access upon request to the vendor. However, currently, the tool requires paid subscription with a fifteen-day free trial option.
6. Discussion
The case studies mentioned here substantiate the target disease prediction accuracy for a drug by this protocol. The finding that imipramine could be repurposed to treat small-cell lung cancer was an expected finding as transcriptomics data analysis was employed by both the literature described methods, as well as by this in silico protocol. However, the correct prediction that the dry form of AMD may be a target disease is noteworthy because the underlying discovery methods were different. The literature [
6] used a small molecule screening technique, while this in silico protocol applied transcriptomics data analysis. This finding further adds credence to the strategy adopted by this protocol to predict diseases a given drug can be repurposed toward.
The prediction that imipramine could be repurposed against SARS is a significant observation. SARS is a deadly viral disease, and during the period of 2002 to 2003, an outbreak caused over 8000 cases with 772 deaths reported in 37 countries [
25]. As of today, there is no treatment or vaccines available for SARS. This makes the predictions made by this protocol particularly advantageous for such deadly diseases by giving patients a possible treatment [
26]. Because this is a novel finding, further clinical investigation measuring the potency of imipramine against SARS in human would be needed.
The in silico protocol described in this study leverages pre-processed transcriptomics data available through the BaseSpace Correlation Engine database. This method is straight forward, simple to use and does not require advanced computer skills. Hence, it may be beneficial to users including researchers, clinicians, patients and even high school students who can apply this method and identify the existing drugs that can be repurposed to serve as new therapeutic targets for various diseases in the future. We hope this protocol could be utilized as a crowdsourcing tool for drug discovery research.
7. Citizen Science
This work describes a protocol by which existing tools and databases can be utilized to draw novel results through human computation. We hope that in the event of epidemics, such as viral infections or transmittable diseases, the global scientific community, especially high school and undergraduate students, could employ this protocol to shortlist drugs that may be useful. We created an online form where the outcomes of such citizen science could be collected. The online form is available at
http://severus.dbmi.pitt.edu/SSDR. Upon receiving submissions with this form, we will post them for world-wide users to view them on a separate page under the same website.