Growth and Morphogenesis during Early Heart Development in Amniotes
Abstract
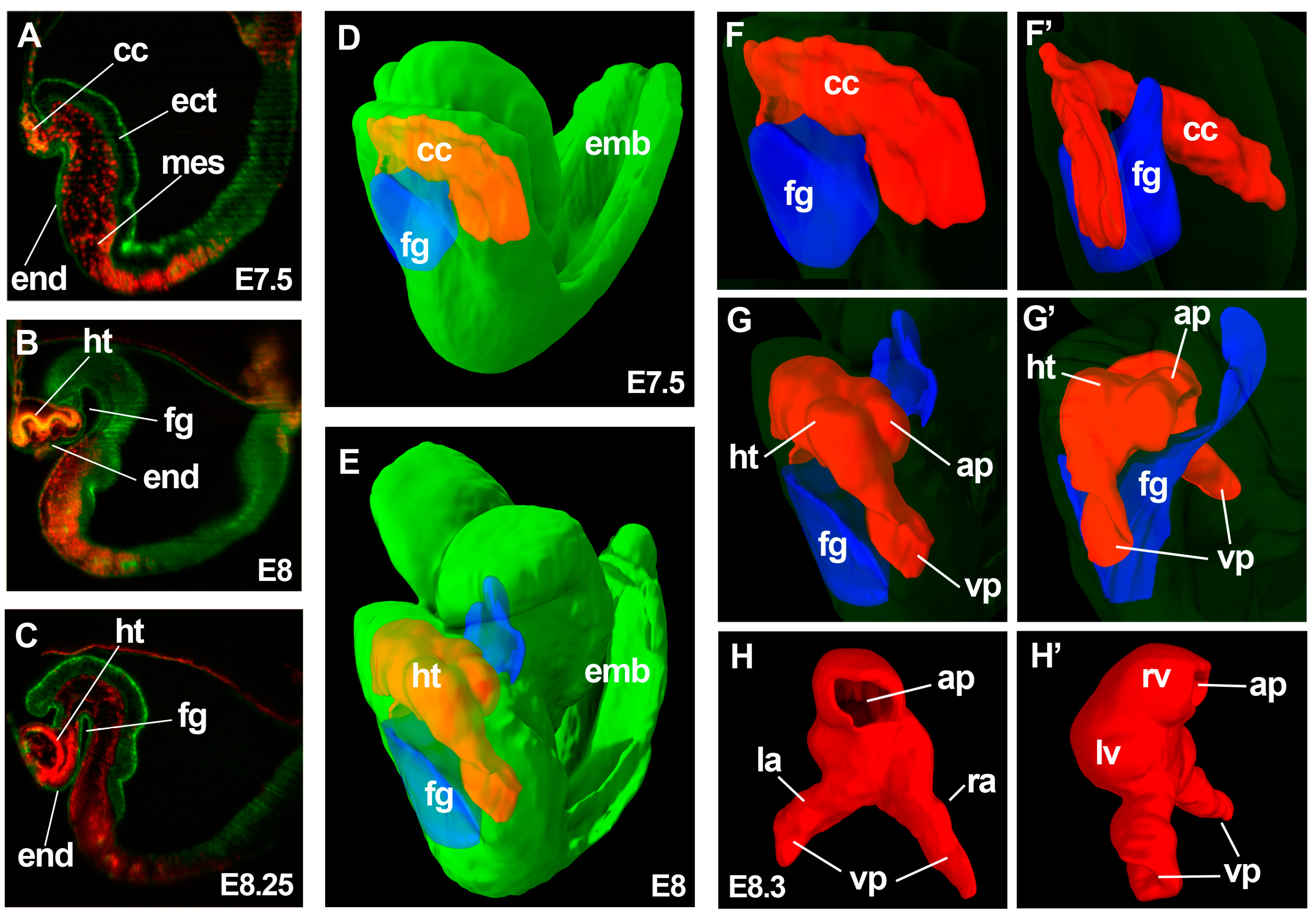
:1. Early Cardiac Development
2. Antero-Posterior Patterning of the Primitive Heart Tube
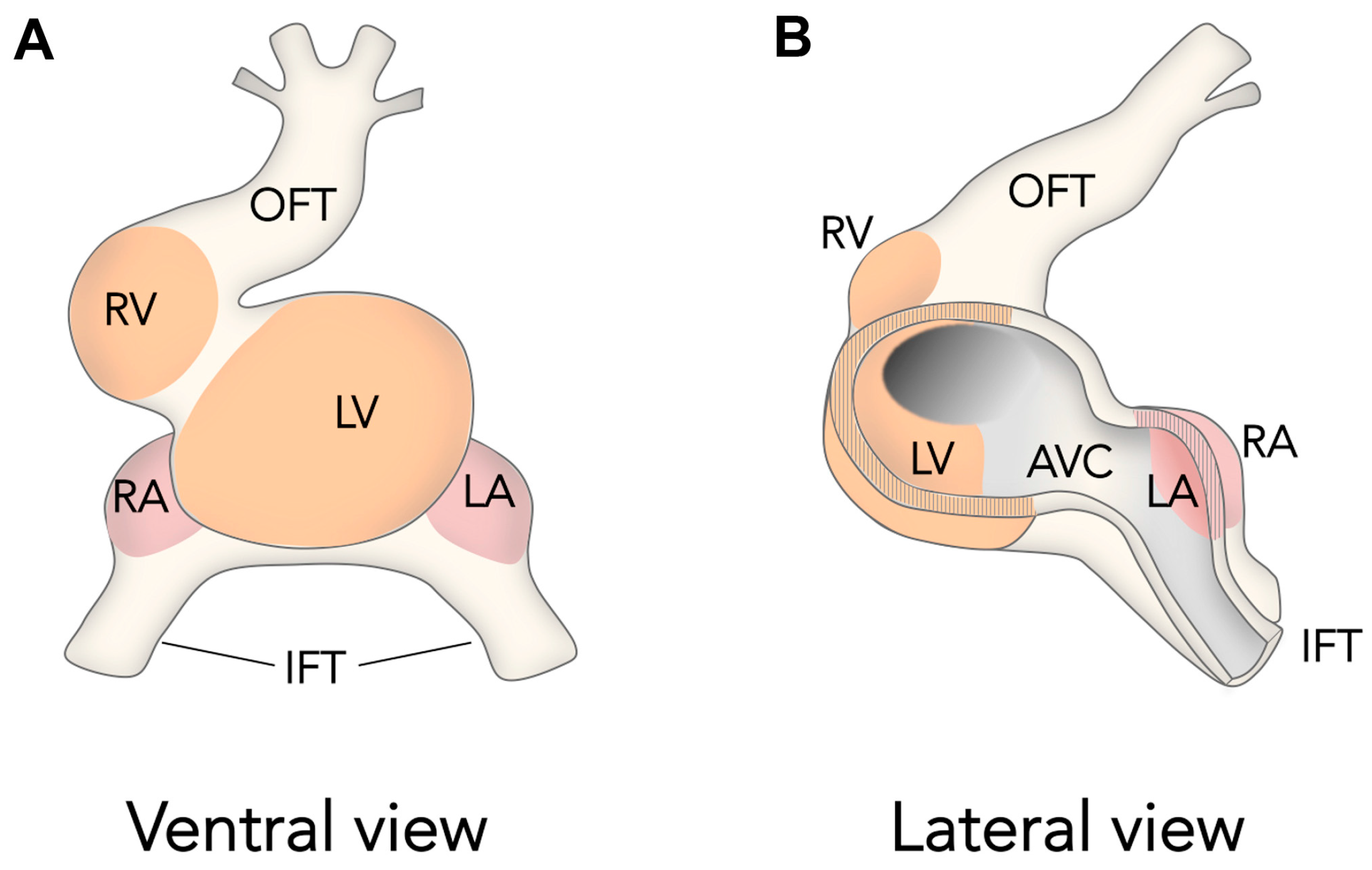
3. Chamber Formation
4. Towards New Quantitative and Dynamic Approaches to Understanding Heart Development
Acknowledgments
Conflicts of Interest
References
- Kirby, M.L. Cardiac Development, 1st ed.; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Rana, M.S.; Christoffels, V.M.; Moorman, A.F. A molecular and genetic outline of cardiac morphogenesis. Acta Physiol. 2013, 207, 588–615. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. 2005, 6, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.G.; Brown, N.A.; Buckingham, M.E. The arterial pole of the mouse heart forms from fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell 2001, 1, 435–440. [Google Scholar] [CrossRef]
- Waldo, K.L.; Kumiski, D.H.; Wallis, K.T.; Stadt, H.A.; Hutson, M.R.; Platt, D.H.; Kirby, M.L. Conotruncal myocardium arises from a secondary heart field. Development 2001, 128, 3179–3188. [Google Scholar] [PubMed]
- Mjaatvedt, C.H.; Nakaoka, T.; Moreno-Rodriguez, R.; Norris, R.A.; Kern, M.J.; Eisenberg, C.A.; Turner, D.; Markwald, R.R. The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 2001, 238, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.L.; Liang, X.; Shi, Y.; Chu, P.H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Galli, D.; Dominguez, J.N.; Zaffran, S.; Munk, A.; Brown, N.A.; Buckingham, M.E. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as pitx2c is expressed. Development 2008, 135, 1157–1167. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, G.; Abu-Issa, R.; de Boer, B.A.; Hutson, M.R.; de Boer, P.A.; Soufan, A.T.; Ruijter, J.M.; Kirby, M.L.; van den Hoff, M.J.; Moorman, A.F. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ. Res. 2009, 104, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Prall, O.W.; Menon, M.K.; Solloway, M.J.; Watanabe, Y.; Zaffran, S.; Bajolle, F.; Biben, C.; McBride, J.J.; Robertson, B.R.; Chaulet, H.; et al. An nkx2-5/bmp2/smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell 2007, 128, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Li, D.; Gupta, M.; Manderfield, L.J.; Ifkovits, J.L.; Wang, Q.; Liu, F.; Liu, Y.; Poleshko, A.; Padmanabhan, A.; et al. Heart development. Integration of bmp and wnt signaling by hopx specifies commitment of cardiomyoblasts. Science 2015, 348. [Google Scholar] [CrossRef] [PubMed]
- Laugwitz, K.L.; Moretti, A.; Caron, L.; Nakano, A.; Chien, K.R. Islet1 cardiovascular progenitors: A single source for heart lineages? Development 2008, 135, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor gata4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.M.; Black, B.L. Mef2c is a direct transcriptional target of isl1 and gata factors in the anterior heart field during mouse embryonic development. Development 2004, 131, 3931–3942. [Google Scholar] [CrossRef] [PubMed]
- Devine, W.P.; Wythe, J.D.; George, M.; Koshiba-Takeuchi, K.; Bruneau, B.G. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Tsuchihashi, T.; Maeda, J.; Shin, C.H.; Ivey, K.N.; Black, B.L.; Olson, E.N.; Yamagishi, H.; Srivastava, D. Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev. Biol. 2011, 351, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Greulich, F.; Rudat, C.; Kispert, A. Mechanisms of t-box gene function in the developing heart. Cardiovasc. Res. 2011, 91, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Mommersteeg, M.T.; Dominguez, J.N.; Wiese, C.; Norden, J.; de Gier-de Vries, C.; Burch, J.B.; Kispert, A.; Brown, N.A.; Moorman, A.F.; Christoffels, V.M. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc. Res. 2010, 87, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.D.; Buckingham, M.E. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 2010, 90, 1–41. [Google Scholar] [PubMed]
- Bruneau, B.G.; Srivastava, D. Congenital heart disease: Entering a new era of human genetics. Circ. Res. 2014, 114, 598–599. [Google Scholar] [CrossRef] [PubMed]
- McCulley, D.J.; Black, B.L. Transcription factor pathways and congenital heart disease. Curr. Top. Dev. Biol. 2012, 100, 253–277. [Google Scholar] [PubMed]
- Merscher, S.; Funke, B.; Epstein, J.A.; Heyer, J.; Puech, A.; Lu, M.M.; Xavier, R.J.; Demay, M.B.; Russell, R.G.; Factor, S.; et al. Tbx1 is responsible for cardiovascular defects in velo-cardio-facial/digeorge syndrome. Cell 2001, 104, 619–629. [Google Scholar] [CrossRef]
- Sylva, M.; van den Hoff, M.J.; Moorman, A.F. Development of the human heart. Am. J. Med. Genet. Part A 2014, 164A, 1347–1371. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Rodriguez, R.A.; Krug, E.L.; Reyes, L.; Villavicencio, L.; Mjaatvedt, C.H.; Markwald, R.R. Bidirectional fusion of the heart-forming fields in the developing chick embryo. Dev. Dyn. 2006, 235, 191–202. [Google Scholar] [CrossRef] [PubMed]
- De Bakker, B.S.; de Jong, K.H.; Hagoort, J.; de Bree, K.; Besselink, C.T.; de Kanter, F.E.; Veldhuis, T.; Bais, B.; Schildmeijer, R.; Ruijter, J.M.; et al. An interactive three-dimensional digital atlas and quantitative database of human development. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. Gata4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, D.; Lu, M.M.; Morrisey, E.E. Advanced cardiac morphogenesis does not require heart tube fusion. Science 2004, 305, 1619–1622. [Google Scholar] [CrossRef] [PubMed]
- George, E.L.; Georges-Labouesse, E.N.; Patel-King, R.S.; Rayburn, H.; Hynes, R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 1993, 119, 1079–1091. [Google Scholar] [PubMed]
- Tyser, R.C.; Miranda, A.M.; Chen, C.M.; Davidson, S.M.; Srinivas, S.; Riley, P.R. Calcium handling precedes cardiac differentiation to initiate the first heartbeat. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- De Boer, B.A.; van den Berg, G.; de Boer, P.A.; Moorman, A.F.; Ruijter, J.M. Growth of the developing mouse heart: An interactive qualitative and quantitative 3d atlas. Dev. Biol. 2012, 368, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Zaffran, S.; Kelly, R.G.; Meilhac, S.M.; Buckingham, M.E.; Brown, N.A. Right ventricular myocardium derives from the anterior heart field. Circ. Res. 2004, 95, 261–268. [Google Scholar] [CrossRef] [PubMed]
- De Jong, F.; Geerts, W.J.; Lamers, W.H.; Los, J.A.; Moorman, A.F. Isomyosin expression pattern during formation of the tubular chicken heart: A three-dimensional immunohistochemical analysis. Anat. Rec. 1990, 226, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.Z.; Bruneau, B.G.; Seidman, J.G.; Seidman, C.E.; Cepko, C.L. Regulation of chamber-specific gene expression in the developing heart by irx4. Science 1999, 283, 1161–1164. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, B.G.; Nemer, G.; Schmitt, J.P.; Charron, F.; Robitaille, L.; Caron, S.; Conner, D.A.; Gessler, M.; Nemer, M.; Seidman, C.E.; et al. A murine model of holt-oram syndrome defines roles of the t-box transcription factor tbx5 in cardiogenesis and disease. Cell 2001, 106, 709–721. [Google Scholar] [CrossRef]
- Mori, A.D.; Zhu, Y.; Vahora, I.; Nieman, B.; Koshiba-Takeuchi, K.; Davidson, L.; Pizard, A.; Seidman, J.G.; Seidman, C.E.; Chen, X.J.; et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 2006, 297, 566–586. [Google Scholar] [CrossRef] [PubMed]
- Basson, C.T.; Bachinsky, D.R.; Lin, R.C.; Levi, T.; Elkins, J.A.; Soults, J.; Grayzel, D.; Kroumpouzou, E.; Traill, T.A.; Leblanc-Straceski, J.; et al. Mutations in human tbx5 [corrected] cause limb and cardiac malformation in holt-oram syndrome. Nat. Genet. 1997, 15, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Y.; Newbury-Ecob, R.A.; Terrett, J.A.; Wilson, D.I.; Curtis, A.R.; Yi, C.H.; Gebuhr, T.; Bullen, P.J.; Robson, S.C.; Strachan, T.; et al. Holt-oram syndrome is caused by mutations in tbx5, a member of the brachyury (t) gene family. Nat. Genet. 1997, 15, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Satin, J.; Fujii, S.; DeHaan, R.L. Development of cardiac beat rate in early chick embryos is regulated by regional cues. Dev. Biol. 1988, 129, 103–113. [Google Scholar] [CrossRef]
- Wu, S.P.; Cheng, C.M.; Lanz, R.B.; Wang, T.; Respress, J.L.; Ather, S.; Chen, W.; Tsai, S.J.; Wehrens, X.H.; Tsai, M.J.; et al. Atrial identity is determined by a coup-tfii regulatory network. Dev. Cell 2013, 25, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Joubin, K.; Stern, C.D. Molecular interactions continuously define the organizer during the cell movements of gastrulation. Cell 1999, 98, 559–571. [Google Scholar] [CrossRef]
- Yutzey, K.E.; Rhee, J.T.; Bader, D. Expression of the atrial-specific myosin heavy chain amhc1 and the establishment of anteroposterior polarity in the developing chicken heart. Development 1994, 120, 871–883. [Google Scholar] [PubMed]
- Hochgreb, T.; Linhares, V.L.; Menezes, D.C.; Sampaio, A.C.; Yan, C.Y.; Cardoso, W.V.; Rosenthal, N.; Xavier-Neto, J. A caudorostral wave of raldh2 conveys anteroposterior information to the cardiac field. Development 2003, 130, 5363–5374. [Google Scholar] [CrossRef] [PubMed]
- Heine, U.I.; Roberts, A.B.; Munoz, E.F.; Roche, N.S.; Sporn, M.B. Effects of retinoid deficiency on the development of the heart and vascular system of the quail embryo. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 1985, 50, 135–152. [Google Scholar] [CrossRef]
- Anderson, C.; Khan, M.A.; Wong, F.; Solovieva, T.; Oliveira, N.M.; Baldock, R.A.; Tickle, C.; Burt, D.W.; Stern, C.D. A strategy to discover new organizers identifies a putative heart organizer. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Cheuvront, T.J.; Lansford, R.D.; Moreno-Rodriguez, R.A.; Schultheiss, T.M.; Rongish, B.J. Dynamic positional fate map of the primary heart-forming region. Dev. Biol. 2009, 332, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, C.; Garcia-Masa, N.; Ganan, C.M.; Garcia-Martinez, V. Movement and commitment of primitive streak precardiac cells during cardiogenesis. Int. J. Dev. Biol. 2009, 53, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Redkar, A.; Montgomery, M.; Litvin, J. Fate map of early avian cardiac progenitor cells. Development 2001, 128, 2269–2279. [Google Scholar] [PubMed]
- Garcia-Martinez, V.; Schoenwolf, G.C. Primitive-streak origin of the cardiovascular system in avian embryos. Dev. Biol. 1993, 159, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, G.C. Location and movements of cardiogenic cells in the chick embryo: The heart-forming portion of the primitive streak. Dev. Biol. 1970, 22, 461–475. [Google Scholar] [CrossRef]
- Kinder, S.J.; Loebel, D.A.; Tam, P.P. Allocation and early differentiation of cardiovascular progenitors in the mouse embryo. Trends Cardiovasc. Med. 2001, 11, 177–184. [Google Scholar] [CrossRef]
- Tam, P.P.; Parameswaran, M.; Kinder, S.J.; Weinberger, R.P. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: The role of ingression and tissue movement during gastrulation. Development 1997, 124, 1631–1642. [Google Scholar] [PubMed]
- Bardot, E.; Calderon, D.; Santoriello, F.; Han, S.; Cheung, K.; Jadhav, B.; Burtscher, I.; Artap, S.; Jain, R.; Epstein, J.; et al. Foxa2 identifies a cardiac progenitor population with ventricular differentiation potential. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Kirby, M.L.; Gale, T.F.; Stewart, D.E. Neural crest cells contribute to normal aorticopulmonary septation. Science 1983, 220, 1059–1061. [Google Scholar] [CrossRef] [PubMed]
- Christoffels, V.M.; Habets, P.E.; Franco, D.; Campione, M.; de Jong, F.; Lamers, W.H.; Bao, Z.Z.; Palmer, S.; Biben, C.; Harvey, R.P.; et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000, 223, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Soufan, A.T.; Ruijter, J.M.; van den Hoff, M.J.; de Boer, P.A.; Hagoort, J.; Moorman, A.F. Three-dimensional reconstruction of gene expression patterns during cardiac development. Physiol. Genom. 2003, 13, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Hoogaars, W.M.; Barnett, P.; Grieskamp, T.; Rana, M.S.; Buermans, H.; Farin, H.F.; Petry, M.; Heallen, T.; Martin, J.F.; et al. Tbx2 and tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell. Mol. Life Sci. 2012, 69, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.L.; Boink, G.J.; Boukens, B.J.; Verkerk, A.O.; van den Boogaard, M.; den Haan, A.D.; Hoogaars, W.M.; Buermans, H.P.; de Bakker, J.M.; Seppen, J.; et al. T-box transcription factor tbx3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res. 2012, 94, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Horsthuis, T.; Farin, H.F.; Grieskamp, T.; Norden, J.; Petry, M.; Wakker, V.; Moorman, A.F.; Christoffels, V.M.; Kispert, A. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ. Res. 2009, 105, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.L.; Boukens, B.J.; Mommersteeg, M.T.; Brons, J.F.; Wakker, V.; Moorman, A.F.; Christoffels, V.M. Transcription factor tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 2008, 102, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, B.G. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- De Boer, B.A.; van den Berg, G.; Soufan, A.T.; de Boer, P.A.; Hagoort, J.; van den Hoff, M.J.; Moorman, A.F.; Ruijter, J.M. Measurement and 3d-visualization of cell-cycle length using double labelling with two thymidine analogues applied in early heart development. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Sizarov, A.; Ya, J.; de Boer, B.A.; Lamers, W.H.; Christoffels, V.M.; Moorman, A.F. Formation of the building plan of the human heart: Morphogenesis, growth, and differentiation. Circulation 2011, 123, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Soufan, A.T.; van den Berg, G.; Ruijter, J.M.; de Boer, P.A.; van den Hoff, M.J.; Moorman, A.F. Regionalized sequence of myocardial cell growth and proliferation characterizes early chamber formation. Circ. Res. 2006, 99, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Christoffels, V.M.; Burch, J.B.; Moorman, A.F. Architectural plan for the heart: Early patterning and delineation of the chambers and the nodes. Trends Cardiovasc. Med. 2004, 14, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Poss, K.D. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 2012, 484, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Person, A.D.; Klewer, S.E.; Runyan, R.B. Cell biology of cardiac cushion development. Int. Rev. Cytol. 2005, 243, 287–335. [Google Scholar] [PubMed]
- Verveer, P.J.; Swoger, J.; Pampaloni, F.; Greger, K.; Marcello, M.; Stelzer, E.H. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat. Methods 2007, 4, 311–313. [Google Scholar] [CrossRef] [PubMed]
- Huisken, J.; Swoger, J.; Del Bene, F.; Wittbrodt, J.; Stelzer, E.H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 2004, 305, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Huisken, J.; Stainier, D.Y. Selective plane illumination microscopy techniques in developmental biology. Development 2009, 136, 1963–1975. [Google Scholar] [CrossRef] [PubMed]
- Mohun, T.J.; Weninger, W.J. Imaging heart development using high-resolution episcopic microscopy. Curr. Opin. Genet. Dev. 2011, 21, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.S.; Lichtman, J.W. Clarifying tissue clearing. Cell 2015, 162, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Fei, P.; Sevag Packard, R.R.; Kang, H.; Xu, H.; Baek, K.I.; Jen, N.; Chen, J.; Yen, H.; Kuo, C.C.; et al. 4-dimensional light-sheet microscopy to elucidate shear stress modulation of cardiac trabeculation. J. Clin. Investig. 2016, 126, 3158. [Google Scholar] [CrossRef] [PubMed]
- Mohun, T.J.; Weninger, W.J. Generation of volume data by episcopic three-dimensional imaging of embryos. Cold Spring Harb. Protoc. 2012, 2012, 681–682. [Google Scholar] [CrossRef] [PubMed]
- Meilhac, S.M.; Lescroart, F.; Blanpain, C.; Buckingham, M.E. Cardiac cell lineages that form the heart. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.E.; Meilhac, S.M. Tracing cells for tracking cell lineage and clonal behavior. Dev. Cell 2011, 21, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Lescroart, F.; Mohun, T.; Meilhac, S.M.; Bennett, M.; Buckingham, M. Lineage tree for the venous pole of the heart: Clonal analysis clarifies controversial genealogy based on genetic tracing. Circ. Res. 2012, 111, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Lescroart, F.; Kelly, R.G.; Le Garrec, J.F.; Nicolas, J.F.; Meilhac, S.M.; Buckingham, M. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development 2010, 137, 3269–3279. [Google Scholar] [CrossRef] [PubMed]
- Meilhac, S.M.; Esner, M.; Kerszberg, M.; Moss, J.E.; Buckingham, M.E. Oriented clonal cell growth in the developing mouse myocardium underlies cardiac morphogenesis. J. Cell Biol. 2004, 164, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Meilhac, S.M.; Esner, M.; Kelly, R.G.; Nicolas, J.F.; Buckingham, M.E. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell 2004, 6, 685–698. [Google Scholar] [CrossRef]
- Meilhac, S.M.; Kelly, R.G.; Rocancourt, D.; Eloy-Trinquet, S.; Nicolas, J.F.; Buckingham, M.E. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development 2003, 130, 3877–3889. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, T.; Fischman, D.A. Retroviral analysis of cardiac morphogenesis: Discontinuous formation of coronary vessels. Proc. Natl. Acad. Sci. USA 1992, 89, 9504–9508. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, T.; Cohen-Gould, L.; Fischman, D.A. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus. Iii: Polyclonal origin of adjacent ventricular myocytes. Dev. Dyn. 1992, 195, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, T.; Borisov, A.; Brown, A.M.; Fischman, D.A. Clonal analysis of cardiac morphogenesis in the chicken embryo using a replication-defective retrovirus: I. Formation of the ventricular myocardium. Dev. Dyn. 1992, 193, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Mikawa, T.; Fischman, D.A.; Dougherty, J.P.; Brown, A.M. In vivo analysis of a new lacz retrovirus vector suitable for cell lineage marking in avian and other species. Exp. Cell Res. 1991, 195, 516–523. [Google Scholar] [CrossRef]
- Lescroart, F.; Chabab, S.; Lin, X.; Rulands, S.; Paulissen, C.; Rodolosse, A.; Auer, H.; Achouri, Y.; Dubois, C.; Bondue, A.; et al. Early lineage restriction in temporally distinct populations of mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 2014, 16, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Preussner, J.; Guenther, S.; Yuan, X.; Yekelchyk, M.; Kuenne, C.; Looso, M.; Zhou, Y.; Braun, T. Single-cell transcriptional regulations and accessible chromatin landscape of cell fate decisions in early heart development. bioRxiv 2017. [Google Scholar] [CrossRef]
- Le Garrec, J.F.; Ragni, C.V.; Pop, S.; Dufour, A.; Olivo-Marin, J.C.; Buckingham, M.E.; Meilhac, S.M. Quantitative analysis of polarity in 3d reveals local cell coordination in the embryonic mouse heart. Development 2013, 140, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, S.; Rodriguez, T.; Clements, M.; Smith, J.C.; Beddington, R.S. Active cell migration drives the unilateral movements of the anterior visceral endoderm. Development 2004, 131, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Hadjantonakis, A.K.; Pisano, E.; Papaioannou, V.E. Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Nowotschin, S.; Hadjantonakis, A.K. Live imaging mouse embryonic development: Seeing is believing and revealing. Methods Mol. Biol. 2014, 1092, 405–420. [Google Scholar] [PubMed]
- Nonaka, S.; Shiratori, H.; Saijoh, Y.; Hamada, H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature 2002, 418, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Udan, R.S.; Piazza, V.G.; Hsu, C.W.; Hadjantonakis, A.K.; Dickinson, M.E. Quantitative imaging of cell dynamics in mouse embryos using light-sheet microscopy. Development 2014, 141, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Nakazato, K.; Keller, P.J.; Kajiura-Kobayashi, H.; Stelzer, E.H.; Mochizuki, A.; Nonaka, S. Live imaging and quantitative analysis of gastrulation in mouse embryos using light-sheet microscopy and 3d tracking tools. Nat. Protoc. 2014, 9, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Nakazato, K.; Keller, P.J.; Kajiura-Kobayashi, H.; Stelzer, E.H.; Mochizuki, A.; Nonaka, S. Live imaging of whole mouse embryos during gastrulation: Migration analyses of epiblast and mesodermal cells. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Ivanovitch, K.; Temino, S.; Torres, M. Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis. bioRxiv 2017. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanovitch, K.; Esteban, I.; Torres, M. Growth and Morphogenesis during Early Heart Development in Amniotes. J. Cardiovasc. Dev. Dis. 2017, 4, 20. https://doi.org/10.3390/jcdd4040020
Ivanovitch K, Esteban I, Torres M. Growth and Morphogenesis during Early Heart Development in Amniotes. Journal of Cardiovascular Development and Disease. 2017; 4(4):20. https://doi.org/10.3390/jcdd4040020
Chicago/Turabian StyleIvanovitch, Kenzo, Isaac Esteban, and Miguel Torres. 2017. "Growth and Morphogenesis during Early Heart Development in Amniotes" Journal of Cardiovascular Development and Disease 4, no. 4: 20. https://doi.org/10.3390/jcdd4040020




