Multi-Scale Assessments of Cardiac Electrophysiology Reveal Regional Heterogeneity in Health and Disease
Abstract
:1. Introduction
2. Developmental Signals Regulating LV-RV Cardiomyocyte Programming
3. Diseases Associated with a Ventricular Chamber Predilection
4. Techniques to Study Regional Electrophysiological Differences
4.1. Multi-Scale Assessment of Cardiac Electrical Properties
4.2. Isolated Cardiomyocytes
4.3. Organotypic Cardiac Slices
4.4. Cardiac Wedge Preparations
4.5. Intact Whole Hearts
4.6. Sharp Microelectrode Intracellular Recordings on Intact Cardiac Tissue
4.7. Optical Mapping
4.8. Optogenetics
4.9. Electrocardiographic Imaging (ECGI)
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Li, X.; Martinez-Fernandez, A.; Hartjes, K.A.; Kocher, J.P.; Olson, T.M.; Terzic, A.; Nelson, T.J. Transcriptional atlas of cardiogenesis maps congenital heart disease interactome. Physiol. Genom. 2014, 46, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Sivagangabalan, G.; Nazzari, H.; Bignolais, O.; Maguy, A.; Naud, P.; Farid, T.; Masse, S.; Gaborit, N.; Varro, A.; Nair, K.; et al. Regional ion channel gene expression heterogeneity and ventricular fibrillation dynamics in human hearts. PLoS ONE 2014, 9, e82179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, R.M.; Firulli, B.A.; Conway, S.J.; Vincentz, J.W.; Firulli, A.B. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev. Dyn. 2010, 239, 3086–3097. [Google Scholar] [CrossRef] [PubMed]
- Rochais, F.; Mesbah, K.; Kelly, R.G. Signaling pathways controlling second heart field development. Circ. Res. 2009, 104, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Molina, C.E.; Heijman, J.; Dobrev, D. Differences in Left versus Right Ventricular Electrophysiological Properties in Cardiac Dysfunction and Arrhythmogenesis. Arrhythm. Electrophysiol. Rev. 2016, 5, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, S.; Zander, J.; Meyers, K.; France, D.; Levine, R.; Porter, G.; Rivkees, S.A.; Morley, G.E.; Fishman, G.I. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc. Natl. Acad. Sci. USA 2002, 99, 10464–10469. [Google Scholar] [CrossRef] [PubMed]
- Rentschler, S.; Yen, A.H.; Lu, J.; Petrenko, N.B.; Lu, M.M.; Manderfield, L.J.; Patel, V.V.; Fishman, G.I.; Epstein, J.A. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation 2012, 126, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Khandekar, A.; Springer, S.; Wang, W.; Hicks, S.; Weinheimer, C.; Diaz-Trelles, R.; Nerbonne, J.M.; Rentschler, S. Notch-Mediated Epigenetic Regulation of Voltage-Gated Potassium Currents. Circ. Res. 2016, 119, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Stroud, D.M.; Darrow, B.J.; Kim, S.D.; Zhang, J.; Jongbloed, M.R.; Rentschler, S.; Moskowitz, I.P.; Seidman, J.; Fishman, G.I. Complex genomic rearrangement in CCS-LacZ transgenic mice. Genesis 2007, 45, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Bezzina, C.R.; Barc, J.; Mizusawa, Y.; Remme, C.A.; Gourraud, J.B.; Simonet, F.; Verkerk, A.O.; Schwartz, P.J.; Crotti, L.; Dagradi, F.; et al. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat. Genet. 2013, 45, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Baruteau, A.E.; Abrams, D.J.; Ho, S.Y.; Thambo, J.B.; McLeod, C.J.; Shah, M.J. Cardiac Conduction System in Congenitally Corrected Transposition of the Great Arteries and Its Clinical Relevance. J. Am. Heart Assoc. 2017, 6, e007759. [Google Scholar] [CrossRef] [PubMed]
- Wallis, G.A.; Debich-Spicer, D.; Anderson, R.H. Congenitally corrected transposition. Orphanet J. Rare Dis. 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.P., Jr.; Bernard, Y.D.; Mellen, B.G.; Celermajer, D.; Baumgartner, H.; Cetta, F.; Connolly, H.M.; Davidson, W.R.; Dellborg, M.; Foster, E.; et al. Long-term outcome in congenitally corrected transposition of the great arteries: A multi-institutional study. J. Am. Coll. Cardiol. 2000, 36, 255–261. [Google Scholar] [CrossRef]
- Saguner, A.M.; Brunckhorst, C.; Duru, F. Arrhythmogenic ventricular cardiomyopathy: A paradigm shift from right to biventricular disease. World J. Cardiol. 2014, 6, 154–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azaouagh, A.; Churzidse, S.; Konorza, T.; Erbel, R. Arrhythmogenic right ventricular cardiomyopathy/dysplasia: A review and update. Clin. Res. Cardiol. 2011, 100, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Sen-Chowdhry, S.; Syrris, P.; Ward, D.; Asimaki, A.; Sevdalis, E.; McKenna, W.J. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation 2007, 115, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Sen-Chowdhry, S.; Morgan, R.D.; Chambers, J.C.; McKenna, W.J. Arrhythmogenic cardiomyopathy: Etiology, diagnosis, and treatment. Ann. Rev. Med. 2010, 61, 233–253. [Google Scholar] [CrossRef] [PubMed]
- Munger, T.M.; Wu, L.Q.; Shen, W.K. Atrial fibrillation. J. Biomed. Res. 2014, 28, 1–17. [Google Scholar] [PubMed]
- Molleman, A. Patch Clamping: An Introductory Guide to Patch Clamp Electrophysiology; John Wiley & Sons: New York, NY, USA, 2003. [Google Scholar]
- Veerman, C.C.; Podliesna, S.; Tadros, R.; Lodder, E.M.; Mengarelli, I.; de Jonge, B.; Beekman, L.; Barc, J.; Wilders, R.; Wilde, A.A.M.; et al. The Brugada Syndrome Susceptibility Gene HEY2 Modulates Cardiac Transmural Ion Channel Patterning and Electrical Heterogeneity. Circ. Res. 2017, 121, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.K.; Springer, S.J.; Wang, W.; Dranoff, E.J.; Zhang, Y.; Kanter, E.M.; Yamada, K.A.; Nerbonne, J.M. Differential Expression and Remodeling of Transient Outward Potassium Currents in Human Left Ventricles. Circ. Arrhythm. Electrophysiol. 2018, 11, e005914. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Franco, J.; Aguilar-Sanchez, Y.; Escobar, A.L. Intact Heart Loose Patch Photolysis Reveals Ionic Current Kinetics During Ventricular Action Potentials. Circ. Res. 2016, 118, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Brandenburger, M.; Wenzel, J.; Bogdan, R.; Richardt, D.; Nguemo, F.; Reppel, M.; Hescheler, J.; Terlau, H.; Dendorfer, A. Organotypic slice culture from human adult ventricular myocardium. Cardiovasc. Res. 2012, 93, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Qiao, Y.; Li, G.; Baechle, K.; Camelliti, P.; Rentschler, S.; Efimov, I.R. Human Organotypic Cultured Cardiac Slices: New Platform For High Throughput Preclinical Human Trials. Sci. Rep. 2016, 6, 28798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watson, S.A.; Scigliano, M.; Bardi, I.; Ascione, R.; Terracciano, C.M.; Perbellini, F. Preparation of viable adult ventricular myocardial slices from large and small mammals. Nat. Protoc. 2017, 12, 2623–2639. [Google Scholar] [CrossRef] [PubMed]
- Semelka, M.; Gera, J.; Usman, S. Sick sinus syndrome: A review. Am. Fam. Physician 2013, 87, 691–696. [Google Scholar]
- Alig, J.; Marger, L.; Mesirca, P.; Ehmke, H.; Mangoni, M.E.; Isbrandt, D. Control of heart rate by cAMP sensitivity of HCN channels. Proc. Natl. Acad. Sci. USA 2009, 106, 12189–12194. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.H.; Burstein, B.; Qi, X.Y.; Sakabe, M.; Chartier, D.; Comtois, P.; Wang, Z.; Kuo, C.T.; Nattel, S. Funny current downregulation and sinus node dysfunction associated with atrial tachyarrhythmia: A molecular basis for tachycardia-bradycardia syndrome. Circulation 2009, 119, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Lipovsky, C.; Hicks, S.; Bhatnagar, S.; Li, G.; Khandekar, A.; Guzy, R.; Woo, K.V.; Nichols, C.G.; Efimov, I.R.; et al. Transient Notch Activation Induces Long-Term Gene Expression Changes Leading to Sick Sinus Syndrome in Mice. Circ. Res. 2017, 121, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.W.; Wang, D.W.; Dyment, M.; Knilans, T.K.; Fish, F.A.; Strieper, M.J.; Rhodes, T.H.; George, A.L., Jr. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J. Clin. Investig. 2003, 112, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Golovko, V.; Gonotkov, M.; Lebedeva, E. Effects of 4-aminopyridine on action potentials generation in mouse sinoauricular node strips. Physiol. Rep. 2015, 3, e12447. [Google Scholar] [CrossRef] [PubMed]
- Sicouri, S.; Belardinelli, L.; Carlsson, L.; Antzelevitch, C. Potent antiarrhythmic effects of chronic amiodarone in canine pulmonary vein sleeve preparations. J. Cardiovasc. Electrophysiol. 2009, 20, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Laughner, J.I.; Ng, F.S.; Sulkin, M.S.; Arthur, R.M.; Efimov, I.R. Processing and analysis of cardiac optical mapping data obtained with potentiometric dyes. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H753–H765. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.S.; Hayashi, H.; Tang, L.; Ogawa, M.; Hernandez, H.; Tan, A.Y.; Li, H.; Karagueuzian, H.S.; Weiss, J.N.; Lin, S.F.; et al. Intracellular calcium and vulnerability to fibrillation and defibrillation in Langendorff-perfused rabbit ventricles. Circulation 2006, 114, 2595–2603. [Google Scholar] [CrossRef] [PubMed]
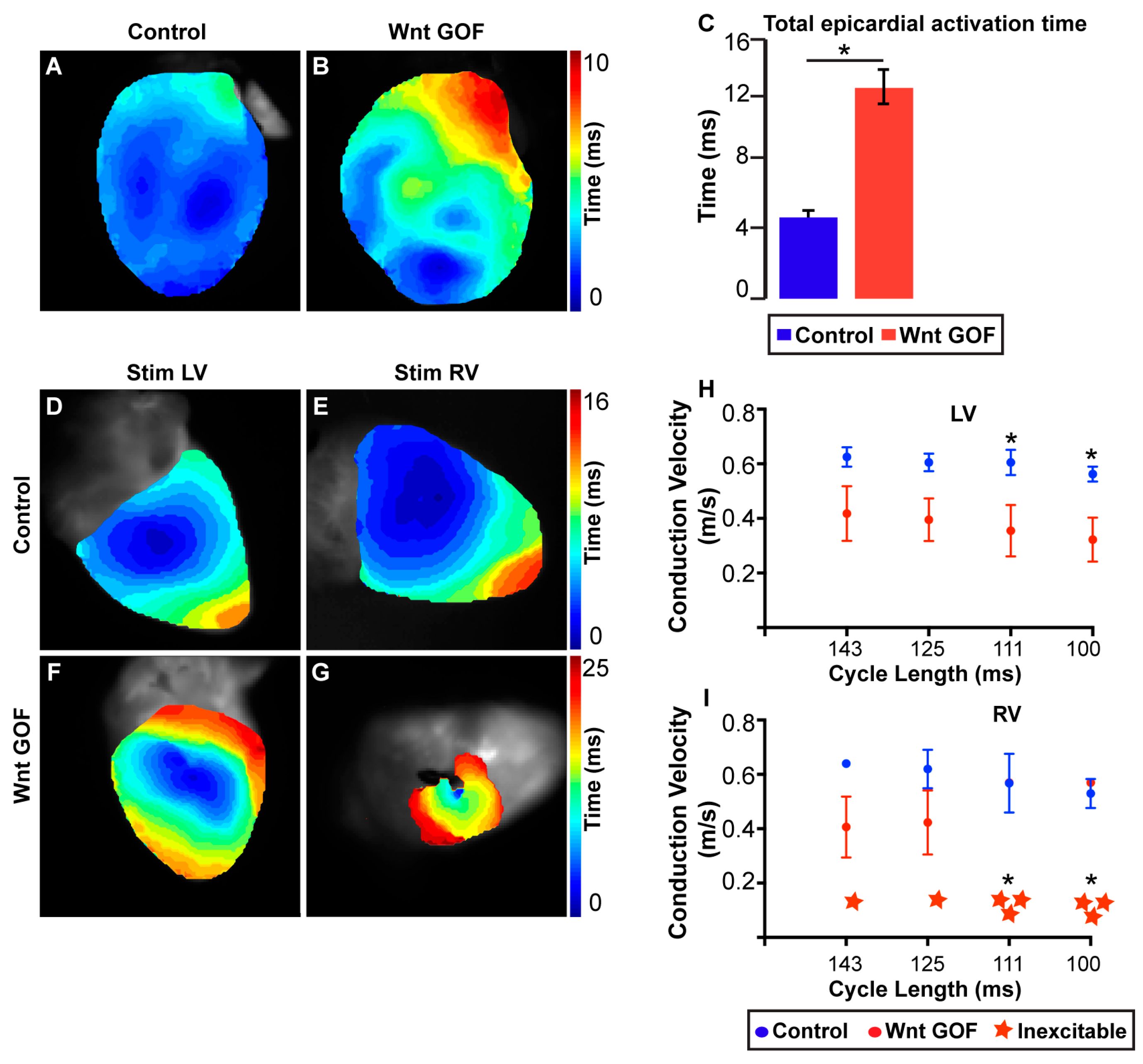
- Gillers, B.S.; Chiplunkar, A.; Aly, H.; Valenta, T.; Basler, K.; Christoffels, V.M.; Efimov, I.R.; Boukens, B.J.; Rentschler, S. Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ. Res. 2015, 116, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Crocini, C.; Coppini, R.; Ferrantini, C.; Yan, P.; Loew, L.M.; Tesi, C.; Cerbai, E.; Poggesi, C.; Pavone, F.S.; Sacconi, L. Defects in T-tubular electrical activity underlie local alterations of calcium release in heart failure. Proc. Natl. Acad. Sci. USA 2014, 111, 15196–15201. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.; Ghouri, I.A.; Kemi, O.J.; Bishop, M.J.; Bernus, O.; Fenton, F.H.; Myles, R.C.; Burton, F.L.; Smith, G.L. Subepicardial action potential characteristics are a function of depth and activation sequence in isolated rabbit hearts. Circ. Arrhythm. Electrophysiol. 2013, 6, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Kotlikoff, M.I. Genetically encoded Ca2+ indicators: Using genetics and molecular design to understand complex physiology. J. Physiol. 2007, 578 Pt 1, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Tallini, Y.N.; Ohkura, M.; Choi, B.R.; Ji, G.; Imoto, K.; Doran, R.; Lee, J.; Plan, P.; Wilson, J.; Xin, H.B.; et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. USA 2006, 103, 4753–4758. [Google Scholar] [CrossRef] [PubMed]
- Guru, A.; Post, R.J.; Ho, Y.Y.; Warden, M.R. Making Sense of Optogenetics. Int. J. Neuropsychopharmacol. 2015, 18. [Google Scholar] [CrossRef] [PubMed]
- Entcheva, E. Cardiac optogenetics. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H1179–H1191. [Google Scholar] [CrossRef] [PubMed]
- Bruegmann, T.; Malan, D.; Hesse, M.; Beiert, T.; Fuegemann, C.J.; Fleischmann, B.K.; Sasse, P. Optogenetic control of heart muscle in vitro and in vivo. Nat. Methods 2010, 7, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Bruegmann, T.; Boyle, P.M.; Vogt, C.C.; Karathanos, T.V.; Arevalo, H.J.; Fleischmann, B.K.; Trayanova, N.A.; Sasse, P. Optogenetic defibrillation terminates ventricular arrhythmia in mouse hearts and human simulations. J. Clin. Investig. 2016, 126, 3894–3904. [Google Scholar] [CrossRef] [PubMed]
- Nyns, E.C.A.; Kip, A.; Bart, C.I.; Plomp, J.J.; Zeppenfeld, K.; Schalij, M.J.; de Vries, A.A.F.; Pijnappels, D.A. Optogenetic termination of ventricular arrhythmias in the whole heart: Towards biological cardiac rhythm management. Eur. Heart J. 2017, 38, 2132–2136. [Google Scholar] [CrossRef] [PubMed]
- Crocini, C.; Ferrantini, C.; Coppini, R.; Scardigli, M.; Yan, P.; Loew, L.M.; Smith, G.; Cerbai, E.; Poggesi, C.; Pavone, F.S.; et al. Optogenetics design of mechanistically-based stimulation patterns for cardiac defibrillation. Sci. Rep. 2016, 6, 35628. [Google Scholar] [CrossRef] [PubMed]
- Rudy, Y. Noninvasive imaging of cardiac electrophysiology and arrhythmia. Ann. N. Y. Acad. Sci. 2010, 1188, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, R.; Silva, J.N.A.; Desouza, K.A.; Abraham, R.L.; Strom, M.; Sacher, F.; Van Hare, G.F.; Haissaguerre, M.; Roden, D.M.; Rudy, Y. Electrophysiologic substrate in congenital Long QT syndrome: Noninvasive mapping with electrocardiographic imaging (ECGI). Circulation 2014, 130, 1936–1943. [Google Scholar] [CrossRef] [PubMed]
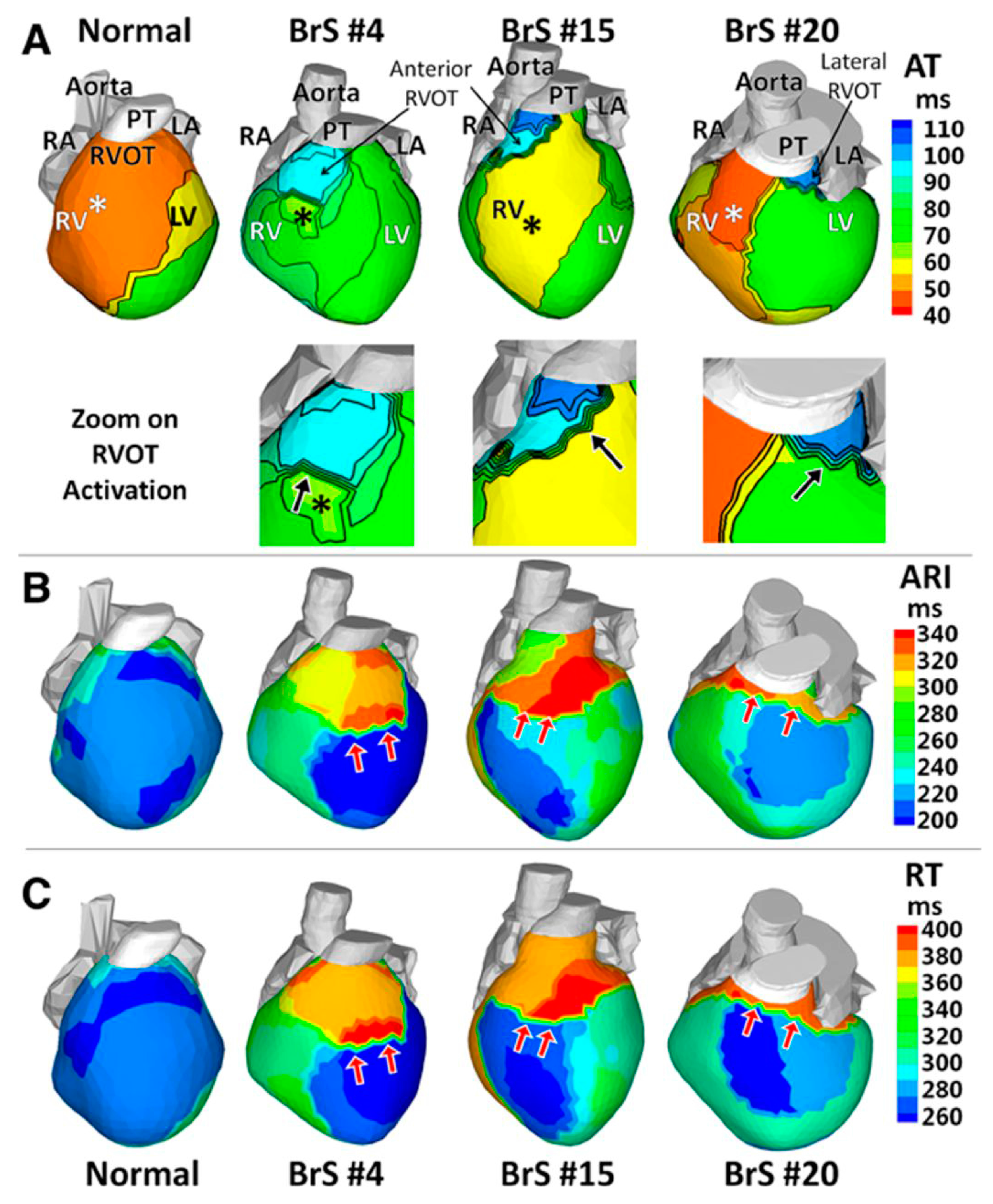
- Zhang, J.; Sacher, F.; Hoffmayer, K.; O’Hara, T.; Strom, M.; Cuculich, P.; Silva, J.; Cooper, D.; Faddis, M.; Hocini, M.; et al. Cardiac electrophysiological substrate underlying the ECG phenotype and electrogram abnormalities in Brugada syndrome patients. Circulation 2015, 131, 1950–1959. [Google Scholar] [CrossRef] [PubMed]
- Cuculich, P.S.; Zhang, J.; Wang, Y.; Desouza, K.A.; Vijayakumar, R.; Woodard, P.K.; Rudy, Y. The electrophysiological cardiac ventricular substrate in patients after myocardial infarction: Noninvasive characterization with electrocardiographic imaging. J. Am. Coll. Cardiol. 2011, 58, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Cuculich, P.S.; Schill, M.R.; Kashani, R.; Mutic, S.; Lang, A.; Cooper, D.; Faddis, M.; Gleva, M.; Noheria, A.; Smith, T.W.; et al. Noninvasive Cardiac Radiation for Ablation of Ventricular Tachycardia. N. Engl. J. Med. 2017, 377, 2325–2336. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lipovsky, C.E.; Brumback, B.D.; Khandekar, A.; Rentschler, S.L. Multi-Scale Assessments of Cardiac Electrophysiology Reveal Regional Heterogeneity in Health and Disease. J. Cardiovasc. Dev. Dis. 2018, 5, 16. https://doi.org/10.3390/jcdd5010016
Lipovsky CE, Brumback BD, Khandekar A, Rentschler SL. Multi-Scale Assessments of Cardiac Electrophysiology Reveal Regional Heterogeneity in Health and Disease. Journal of Cardiovascular Development and Disease. 2018; 5(1):16. https://doi.org/10.3390/jcdd5010016
Chicago/Turabian StyleLipovsky, Catherine E., Brittany D. Brumback, Aditi Khandekar, and Stacey L. Rentschler. 2018. "Multi-Scale Assessments of Cardiac Electrophysiology Reveal Regional Heterogeneity in Health and Disease" Journal of Cardiovascular Development and Disease 5, no. 1: 16. https://doi.org/10.3390/jcdd5010016




