The Role of scaRNAs in Adjusting Alternative mRNA Splicing in Heart Development
Abstract
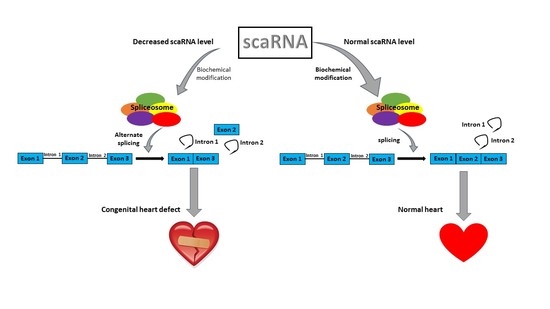
:1. Introduction
2. Material and Methods
2.1. Subjects
2.2. Derivation of Primary Cells
2.3. Transfection of scaRNA Plasmids into Primary Cells
2.4. scaRNA Knockdown
2.5. RNA Isolation and qRT-PCR (Human Tissue)
2.6. Microarray Analysis of Splice Variants
2.7. Zebrafish
2.8. Morpholino Oligonucleotide Injections and Morphological Analysis
2.9. RNA-Seq Analysis
3. Results and Discussion
3.1. Cardiac Regulatory Networks Are Enriched for Alternative Splice Isoforms in TOF
3.2. Primary Cell Lines Derived from TOF Myocardium Retain the Same Relative Expression Patterns as the Tissue
3.3. Overexpression of ACA26 and ACA35 in TOF Primary Cells Was Associated with an Increase in U2 Levels and a Decrease in Fetal Splice Isoforms
3.4. Knockdown of scaRNAs (ACA35 Targeting U2, or U94 Targeting U6)
3.5. Splice Isoforms Change during Development in Zebrafish and after Targeted Knockdown of scaRNAs
4. Conclusions
Funding
Conflicts of Interest
References
- Gelb, B.D.; Chung, W.K. Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb. Perspect. Med. 2014, 4, a013953. [Google Scholar] [CrossRef] [PubMed]
- Ware, S.M.; Jefferies, J.L. New Genetic Insights into Congenital Heart Disease. J. Clin. Exp. Cardiol. 2012, S8. [Google Scholar] [CrossRef]
- Kalsotra, A.; Cooper, T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011, 12, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Bentham, J.; Bhattacharya, S. Genetic mechanisms controlling cardiovascular development. Ann. N. Y. Acad. Sci. 2008, 1123, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, B.G. The developmental genetics of congenital heart disease. Nature 2008, 451, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Meilhac, S.; Zaffran, S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005, 6, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.B.; Liu, Y.L.; Sun, P.W.; Lv, X.D.; Du, M.; Fan, X.M. Molecular mechanisms of congenital heart disease. Cardiovasc. Pathol. 2009, 19, e183–e193. [Google Scholar] [PubMed]
- Thum, T.; Catalucci, D.; Bauersachs, J. MicroRNAs: Novel regulators in cardiac development and disease. Cardiovasc. Res. 2008, 79, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Wessels, M.W.; Willems, P.J. Genetic factors in non-syndromic congenital heart malformations. Clin. Genet. 2010, 78, 103–123. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Wang, K.; Li, P.F.; Cooper, T.A. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010, 24, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Bland, C.S.; Wang, E.T.; Vu, A.; David, M.P.; Castle, J.C.; Johnson, J.M.; Burge, C.B.; Cooper, T.A. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010, 38, 7651–7664. [Google Scholar] [CrossRef] [PubMed]
- Salomonis, N.; Nelson, B.; Vranizan, K.; Pico, A.R.; Hanspers, K.; Kuchinsky, A.; Ta, L.; Mercola, M.; Conklin, B.R. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLoS Comput. Biol. 2009, 5, e1000553. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.J.; Cooper, T.A. The pathobiology of splicing. J. Pathol. 2010, 220, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Cooper, T.A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012, 18, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Kaida, D.; Schneider-Poetsch, T.; Yoshida, M. Splicing in oncogenesis and tumor suppression. Cancer Sci. 2012, 103, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Tanackovic, G.; Ransijn, A.; Thibault, P.; Abou Elela, S.; Klinck, R.; Berson, E.L.; Chabot, B.; Rivolta, C. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 2011, 20, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, K.; Liao, J.; Jiang, F. Small nucleolar RNAs in cancer. Biochim. Biophys. Acta 2012, 1826, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Kalsotra, A.; Xiao, X.; Ward, A.J.; Castle, J.C.; Johnson, J.M.; Burge, C.B.; Cooper, T.A. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. USA 2008, 105, 20333–20338. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Kapranov, P.; Drenkow, J.; Dike, S.; Brubaker, S.; Patel, S.; Long, J.; Stern, D.; Tammana, H.; Helt, G.; et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005, 308, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Olson, E.N. MicroRNA regulatory networks in cardiovascular development. Dev. Cell 2010, 18, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T.; Filipowicz, W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 1995, 9, 1411–1424. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.E., Jr.; Kibiryeva, N.; Zhou, X.G.; Marshall, J.A.; Lofland, G.K.; Artman, M.; Chen, J.; Bittel, D.C. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ. Cardiovasc. Genet. 2012, 5, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Karijolich, J.; Yu, Y.T. Spliceosomal snRNA modifications and their function. RNA Biol. 2010, 7, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Karunatilaka, K.S.; Rueda, D. Post-transcriptional modifications modulate conformational dynamics in human U2-U6 snRNA complex. RNA 2014, 20, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Bittel, D.C.; Butler, M.G.; Kibiryeva, N.; Marshall, J.A.; Chen, J.; Lofland, G.K.; O’Brien, J.E., Jr. Gene expression in cardiac tissues from infants with idiopathic conotruncal defects. BMC Med. Genom. 2011, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Colak, R.; et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Higa-Nakamine, S.; Suzuki, T.; Uechi, T.; Chakraborty, A.; Nakajima, Y.; Nakamura, M.; Hirano, N.; Kenmochi, N. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res. 2012, 40, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Ju, R.; Cirone, P.; Lin, S.; Griesbach, H.; Slusarski, D.C.; Crews, C.M. Activation of the planar cell polarity formin DAAM1 leads to inhibition of endothelial cell proliferation, migration, and angiogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 6906–6911. [Google Scholar] [CrossRef] [PubMed]
- Darzacq, X.; Jady, B.E.; Verheggen, C.; Kiss, A.M.; Bertrand, E.; Kiss, T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002, 21, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Zhou, J.; Kahen, E.; Terns, R.M.; Terns, M.P.; Li, H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat. Struct. Mol. Biol. 2009, 16, 740–746. [Google Scholar] [CrossRef] [PubMed]




| Affy Probe Set ID | Target | Chromosome | Alternate Name | Host Gene |
|---|---|---|---|---|
| HBII-382_s_st | U2 snRNA C61 and U2 snRNA G11 | 1 | ||
| mgU2-19-30 | U2 snRNA G19 and U2 snRNA A30 | X | scaRNA9 | |
| mgU2-25-61_st | U2 snRNA G25 and U2 snRNA C61 | 1 | scaRNA2 | |
| U92_st | U2 snRNA U34 and U2 snRNA U44 | 9 | scaRNA8 | FAM29A |
| ACA26_st | U2 snRNA U41 and U2 snRNA U39 | 1 | scaRNA4 | KIAA0907 |
| ACA35_st | U2 snRNA U89 | 1 | scaRNA1 | PPP1R8 |
| mgU6-47_st | U6 snRNA A47 | 17 | SNORD7 | |
| mgU6-53_st | U6 snRNA A53 | 14 | SNORD8 | CHD8 |
| mgU6-53B_st | U6 snRNA A53 | 14 | SNORD9 | CHD8 |
| HBII-166_st | U6 snRNA C60 | 11 | SNORD67 | CKAP5 |
| U94_st | U6 snRNA C62 | 2 | SNORD94 | PTCD3 |
| ACA12_st | U6 snRNA C62 | X | scaRNA24 | POLA1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagasawa, C.; Ogren, A.; Kibiryeva, N.; Marshall, J.; O’Brien, J.E.; Kenmochi, N.; Bittel, D.C. The Role of scaRNAs in Adjusting Alternative mRNA Splicing in Heart Development. J. Cardiovasc. Dev. Dis. 2018, 5, 26. https://doi.org/10.3390/jcdd5020026
Nagasawa C, Ogren A, Kibiryeva N, Marshall J, O’Brien JE, Kenmochi N, Bittel DC. The Role of scaRNAs in Adjusting Alternative mRNA Splicing in Heart Development. Journal of Cardiovascular Development and Disease. 2018; 5(2):26. https://doi.org/10.3390/jcdd5020026
Chicago/Turabian StyleNagasawa, Chloe, Allison Ogren, Nataliya Kibiryeva, Jennifer Marshall, James E. O’Brien, Naoya Kenmochi, and Douglas C. Bittel. 2018. "The Role of scaRNAs in Adjusting Alternative mRNA Splicing in Heart Development" Journal of Cardiovascular Development and Disease 5, no. 2: 26. https://doi.org/10.3390/jcdd5020026