Phylogenetic Analysis of the Synnema-Producing Genus Synnemapestaloides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. DNA Extraction and Polymerase Chain-Reaction (PCR)
2.3. Molecular Phylogeny
2.4. Morphological Observations
3. Results
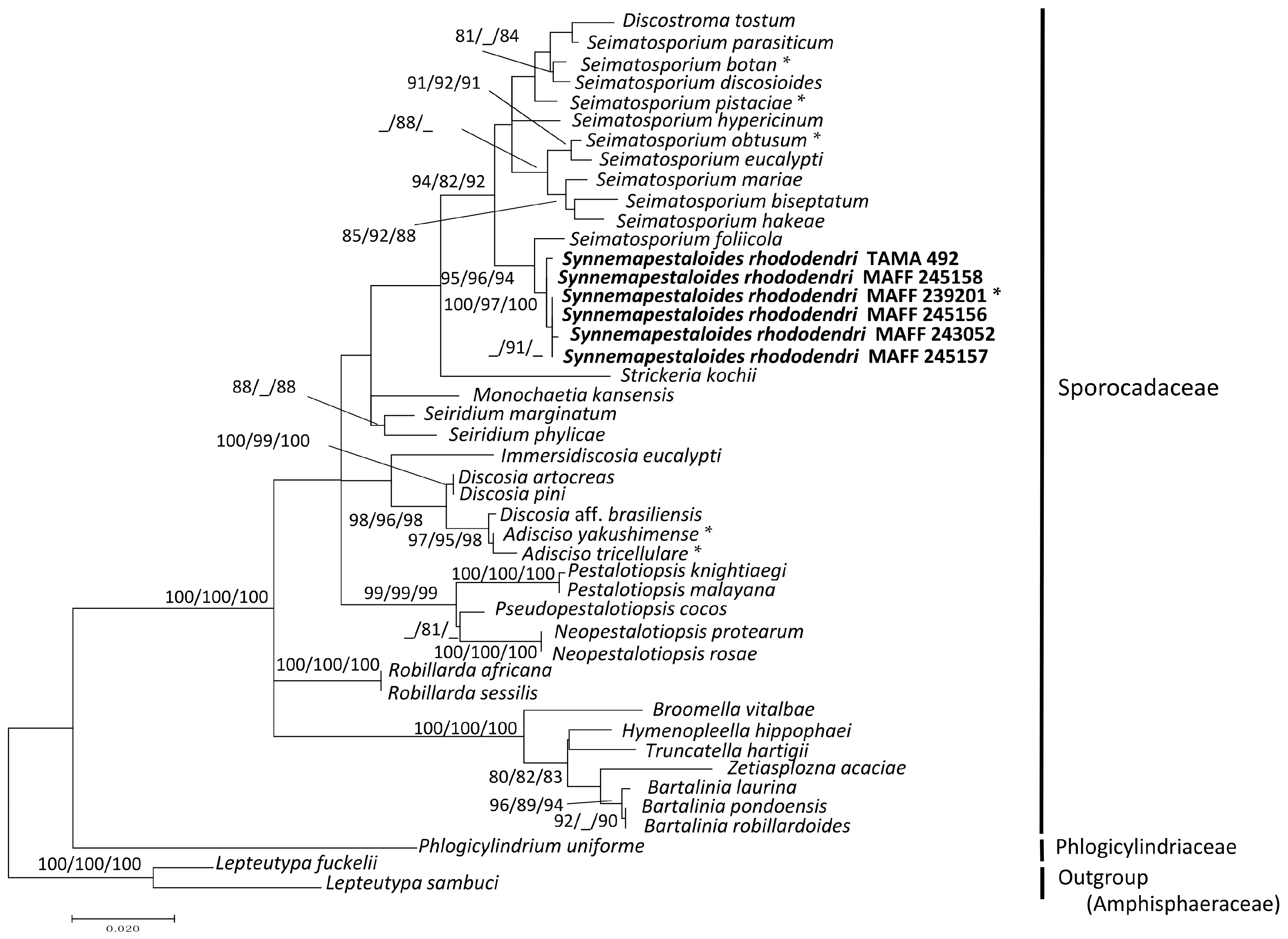
3.1. Phylogenetic Analysis of the ITS and LSU D1–D2 Region
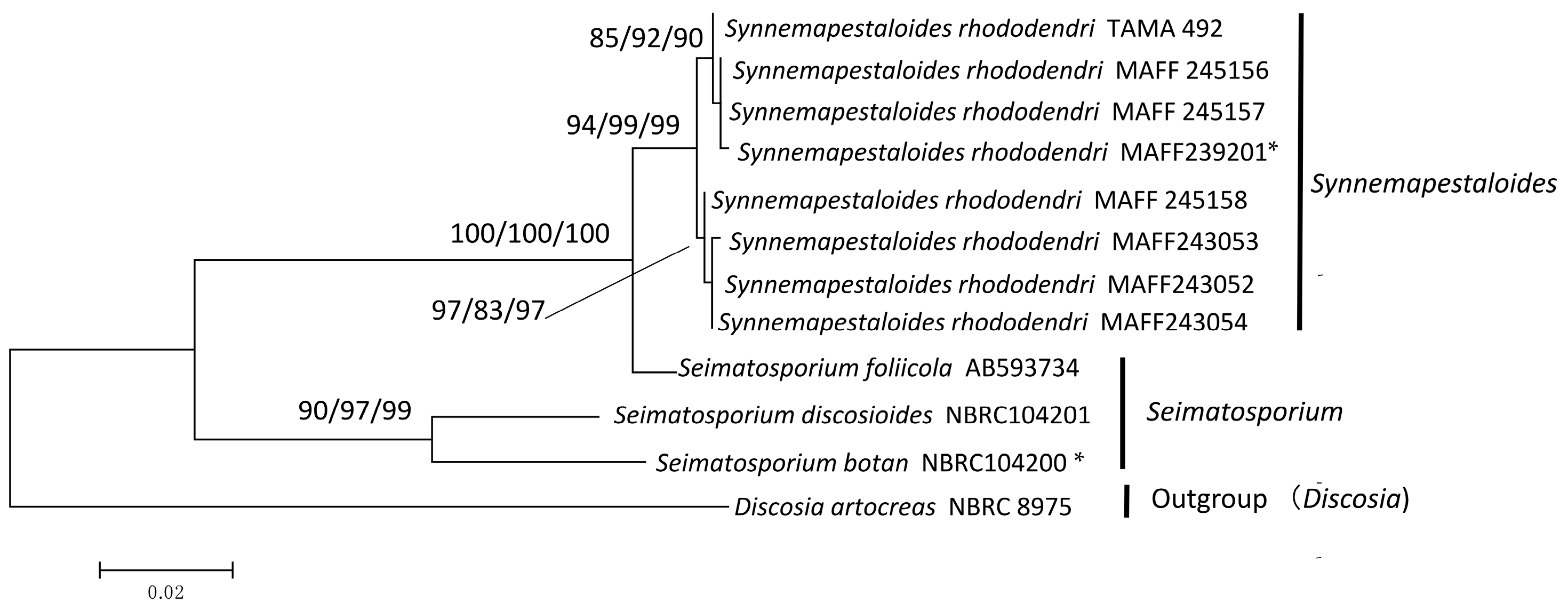
3.2. Phylogenetic Analysis of ITS and β-Tubulin
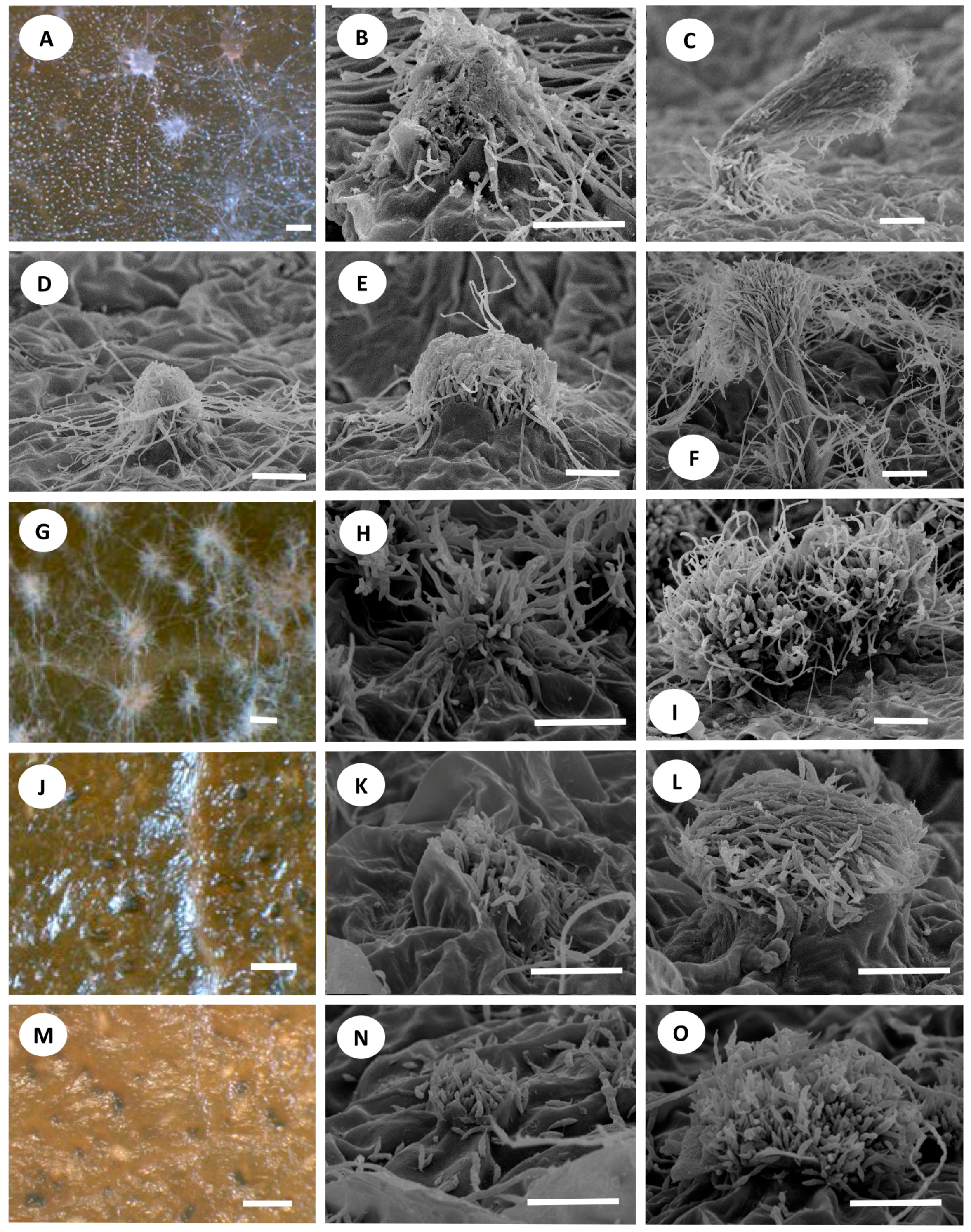
3.3. Development of Conidiomata and Conidial Structures
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix
Taxonomy
Synnemapestaloides T. Handa & Y. Harada emend. Kyoko Watan., Nozawa, Kaz. Tanaka & Toy. Sato
Synnemapestaloides foliiola (Berkeley) Kyoko Watan. Nozawa, Kaz. Tanaka & Toy. Sato comb. nov. [MB818747]
References
- Nag Raj, T.R. Coelomycetous Anamorphs with Appendage Bearing Conidia; Mycologue: Waterloo, ON, Canada, 1993. [Google Scholar]
- Kirk, P.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Ainsworth and Bisby’s Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, ON, Canada, 2008. [Google Scholar]
- Jeewon, R.; Liew, E.C.Y.; Hyde, K.D. Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mycol. Res. 2002, 25, 378–392. [Google Scholar] [CrossRef]
- Jeewon, R.; Liew, E.C.Y.; Hyde, K.D. Molecular systematics of the Amphisphaeriaceae based on cladistic analyses of partial LSU rDNA gene sequences. Mycol. Res. 2003, 107, 1392–1402. [Google Scholar] [CrossRef]
- Lee, S.; Crous, P.W.; Wingfield, M.J.; Sacc, B. Pestalotioid fungi from Restionaceae in the Cape Floral Kingdom. Stud. Mycol. 2006, 55, 175–187. [Google Scholar] [CrossRef]
- Tanaka, K.; Endo, M.; Hirayama, K.; Okane, I.; Hosoya, T.; Sato, T. Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 2011, 26, 85–98. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.B.G.; Mckenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Gardiennet, A.; Voglmayr, H. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 2016, 37, 82–105. [Google Scholar]
- Handa, T.; Fujita, T.; Harada, Y. Synnemapestaloides rhododendri, a new genus and new species of synnematous hyphomycete, causing synnemapestaloides twig blight disease of Rhododendron brachycarpum in Japan. Mycoscience 2004, 45, 137–142. [Google Scholar] [CrossRef]
- De Notaris, G. Micromycetes italiei Dec II. Mem. Accad. Sci. Torino 1839, 3, 80–81. [Google Scholar]
- Hyde, K.D.; McKenzie, E.H.C.; KoKo, T.W. Towards incorporating anamorphic fungi in a natural classification—Checklist and notes for 2010. Mycosphere 2011, 2, 1–88. [Google Scholar]
- O’Donnell, K. Fusarium and its near relatives. In The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CAB International: Wallingford, UK, 1993; pp. 225–233. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. AEM 1995, 61, 1323–1330. [Google Scholar]
- Thompson, J.; Higgins, G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4690. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Watanabe, K.; Doi, Y.; Kobayashi, T. Conidiomatal development of Pestalotiopsis guepinii and P. neglecta on leaves of Gardenia jasminoides. Mycoscience 1998, 39, 71–75. [Google Scholar] [CrossRef]
- Watanabe, K.; Kobayashi, T.; Doi, Y. Conidiomata of Truncatella sp. on different media (in Japanese). Nippon Kingakukai Kaiho 1998, 39, 21–25. [Google Scholar]
- Sutton, B.C. The Coelomycetes: Fungi Imperfecti with Pycnidia, Acervuli and Stromata; Commonwealth Mycological Institute: Kew, UK, 1980. [Google Scholar]
- Steyaert, R.L. Contributions à l’étude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.). Bull. Jard. Bot. Etat. Brux. 1949, 19, 285–354. [Google Scholar] [CrossRef]
- Guba, E.F. Monograph of Pestalotia and Monochaetia; Harvard University Press: Cambridge, UK, 1961. [Google Scholar]
- Hanlin, R.T. Conidioma development in Ophiodothella vaccinii. Mycologia 2003, 95, 506–512. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Maharachchikumbura, S.N.N.; Daranagama, D.A.; Bulgakov, T.S.; Bhat, D.J.; Bahkali, A.H.; Hyde, K.D. Towards a backbone tree for Seimatosporium, with S. physocarpi sp. nov. Mycosphere 2015, 6, 385–400. [Google Scholar]

| Species | Strain | GeneBank Accessions | ||
|---|---|---|---|---|
| ITS | LSU | ß-tublin | ||
| Synnemapestaloides rhododendri | MAFF 245156 1 | LC047755 4 | LC047746 | LC047763 |
| Synnemapestaloides rhododendri | MAFF 245157 | LC047756 | LC047747 | LC047764 |
| Synnemapestaloides rhododendri | MAFF 245158 | LC047754 | LC047745 | LC047762 |
| Synnemapestaloides rhododendri | MAFF 243052 | LC047757 | LC047748 | LC047765 |
| Synnemapestaloides rhododendri | MAFF 243053 | LC047758 | – | LC047766 |
| Synnemapestaloides rhododendri | MAFF 243054 | LC047759 | – | LC047767 |
| Synnemapestaloides rhododendri | TAMA 492 2 | LC047760 | LC047749 | LC047768 |
| Synnemapestaloides rhododendri | MAFF 239201 * | LC047753 | LC047744 | LC047761 |
| Bartalinia laurina | HKUCC 6537 | AF405302 | AF382369 | –5 |
| Bartalinia pondoensis | CMW 31067 | GU291796 | GU291796 | – |
| Bartalinia robillardoides | CBS 122705 | KJ710460 | KJ710438 | – |
| Broomella vitalbae | BV = CBS 140412 | KT949895 | KT949895 | – |
| Discosia aff. pleurochaeta | MAFF24779 | AB594781 | AB593713 | – |
| Discosia artocreas | NBRC 8975 | AB594773 | AB593705 | – |
| Discosia pini | MAFF 410149 | AB594776 | AB593708 | – |
| Discosia pleurochaeta | MAFF 242778 | AB594777 | AB593709 | – |
| Discosia tricellulare | NBRC 32705 * | AB594796 | AB593728 | – |
| Discosia yakushimense | MAFF242774 * | AB594789 | AB593721 | – |
| Discostroma tostum | NBRC 32626 | AB594795 | AB593727 | – |
| Hymenopleella hippophaeicola | LH = CBS 140410 | KT949901 | KT949901 | – |
| Immersidiscosia eucalypti | MAFF 242781 | AB594793 | AB593725 | – |
| Lepteutypa fuckelii | RS = CBS 131707 | KT949902 | KT949902 | – |
| Lepteutypa sambuci | LEF = CBS 140409 | KT949904 | KT949904 | – |
| Monochaetia kansensis | PSHI2004Endo1030 | DQ534044 | DQ534035 | – |
| Neoestalotiopsis protearum | CBS 114178 | JN712498 | JN712564 | – |
| Neopestalotiopsis rosae | CBS 101057 | KM199359 | KM116245 | – |
| Pestalotiopsis knightiae | CBS 114138 | KM199310 | KM116227 | – |
| Pestalotiopsis malayana | CBS 102220 | KM199306 | KM116238 | – |
| Phlogicylindrium uniforme | CBS 131312 | JQ044426 | JQ044445 | – |
| Pseudopestalotiopsis cocos | CBS 272.29 | KM199378 | KM116276 | – |
| Robillarda africana | CBS 122.75 | KR873253 | KR873281 | – |
| Robillarda sessilis | CBS 114312 | KR873256 | KR873284 | – |
| Seimatosporium biseptatum | CPC 13584 | JN871199 | JN871208 | – |
| Seimatosporium foliicola | NBRC 32676 3 | AB593734 | AB594802 | LC047769 |
| Seimatosporium hakeae | NBRC 32678 | AB594804 | AB593736 | – |
| Seimatosporium obtusum | CPC 12935 * | JN871206 | JN871215 | – |
| Seimatosporium pistaciae | CBS 138865 | P004463 | KP004491 | – |
| Seimatosporium botan | NBRC 104200 * | AB594799 | AB593731 | LC047770 |
| Seimatosporium discosioides | NBRC 104201 | AB594800 | AB593732 | LC047771 |
| Seimatosporium eucalypti | CBS 115131 | JN871200 | JN871209 | – |
| Seimatosporium hypericinum | NBRC 32647 | AB594805 | AB593737 | – |
| Seimatosporium mariae | NBRC 32681 | AB594807 | AB593740 | – |
| Seimatosporium parasiticum | NBRC 32682 | AB594805 | AB593741 | – |
| Seiridium marginatum | BLO = CBS 140403 | KT949914 | KT949914 | – |
| Seiridium phylicae | CPC 19965 | KC005787 | KC005809 | – |
| Strickeria kochii | C138 | KT949917 | KT949917 | – |
| Truncatella hartigii | CBS 118148 | DQ278913 | DQ278928 | – |
| Zetiasplozna acaciae | CBS 137994 | KJ869149 | KJ869206 | – |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, K.; Sekiguchi, M.; Sato, T.; Hsiang, T.; Kaneko, S.; Tanaka, K.; Kanda, M.; Fujita, N.; Nozawa, S. Phylogenetic Analysis of the Synnema-Producing Genus Synnemapestaloides. J. Fungi 2016, 2, 28. https://doi.org/10.3390/jof2040028
Watanabe K, Sekiguchi M, Sato T, Hsiang T, Kaneko S, Tanaka K, Kanda M, Fujita N, Nozawa S. Phylogenetic Analysis of the Synnema-Producing Genus Synnemapestaloides. Journal of Fungi. 2016; 2(4):28. https://doi.org/10.3390/jof2040028
Chicago/Turabian StyleWatanabe, Kyoko, Mao Sekiguchi, Toyozo Sato, Tom Hsiang, Shigeru Kaneko, Kazuaki Tanaka, Masaru Kanda, Naoko Fujita, and Shunsuke Nozawa. 2016. "Phylogenetic Analysis of the Synnema-Producing Genus Synnemapestaloides" Journal of Fungi 2, no. 4: 28. https://doi.org/10.3390/jof2040028






