Candida glabrata Biofilms: How Far Have We Come?
Abstract
:1. Introduction
1.1. Biology of Candida glabrata
1.2. Epidemiology and Virulence Factors of Candida glabrata
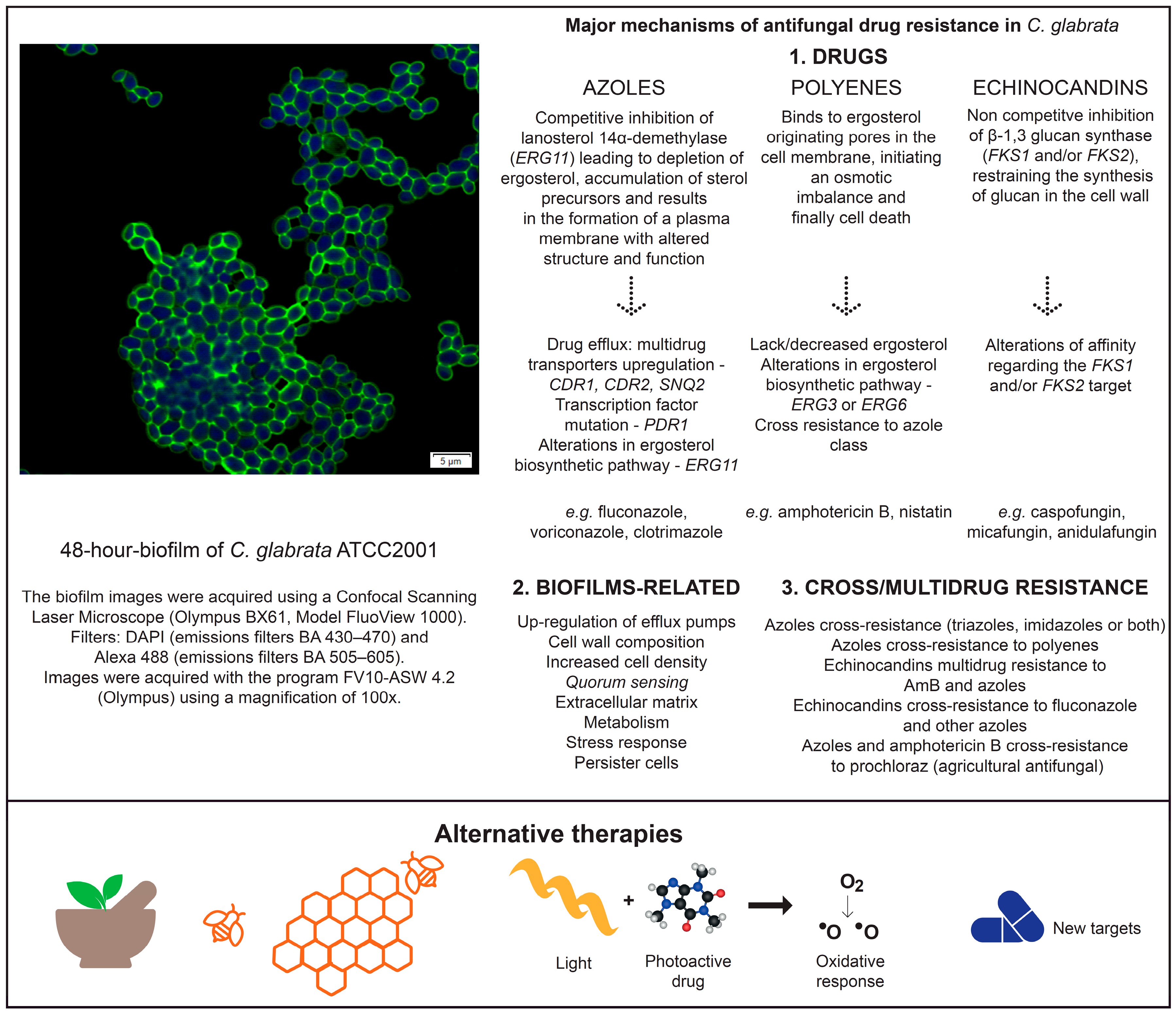
2. General Mechanisms of Antifungal Drug Resistance
2.1. Azole’s Resistance
2.2. Polyene’s Resistance
2.3. Echinocandin’s Resistance
3. Resistance Mechanisms Related to Biofilms
3.1. Up-Regulation of Efflux Pumps and Cell Wall Composition
3.2. Increased Cell Density and Quorum Sensing
3.3. Extracellular Matrix
3.4. Metabolism and Stress Response
3.5. Persister Cells
4. Cross and/or Multidrug Resistance
5. Alternative Therapies for the Treatment of Infections Related to Candida glabrata
5.1. Plant Essential Oils and Extracts
5.2. Honey
5.3. Photodynamic Therapy (PDT)
5.4. Antifungal Agents with New Targets and New Sources
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fidel, P.; Vazquez, J.; Sobel, J. Candida glabrata: Review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 1999, 12, 80–96. [Google Scholar]
- Introduction and historical perspectives. In Candida and Candidiasis; Calderone, R. (Ed.) ASM Press: Washington, DC, USA, 2002; pp. 15–25.
- Larone, H. Medically Important Fungi: A Guide to Identification, 4th ed.; ASM Press: Washington, DC, USA, 2002. [Google Scholar]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Odds, F. Candida and Candidiasis, 2nd ed.; Baillierir Tindall.: London, UK, 1988. [Google Scholar]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Hube, B. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell. Microbiol. 2013, 15, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Domergue, R.; Zupancic, M.L.; Cormack, B.P. A yeast by any other name: Candida glabrata and its interaction with the host. Curr. Opin. Microbiol. 2005, 8, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Hajjeh, R.A.R.A.; Sofair, A.N.A.N.; Harrison, L.H.L.H.; Lyon, G.M.M.; Arthington-Skaggs, B.A.B.A.; Mirza, S.A.S.A.; Phelan, M.; Morgan, J.; Lee-Yang, W.; Ciblak, M.A.M.A.; et al. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J. Clin. Microbiol. 2004, 42, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Silva, S.; Henriques, M. Candida glabrata: A review of its features and resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 673–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, A.L.; Guimaraes, T.; Silva, L.R.B.F.; Monfardini, L.P.; de Almeida Monfardini, L.P.; Cunha, A.K.B.; Rady, P.; Alves, T.; Rosas, R.C. Prospective observational study of candidemia in Sao Paulo, Brazil: Incidence rate, epidemiology, and predictors of mortality. Infect. Control Hosp. Epidemiol. 2007, 28, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E.; Costa, A.; Fasce, R.; Molinari, M.P.; Rosso, R.; Pallavicini, F.B.; Viscoli, C.; Pfaller, M.; Diekema, D.; et al. Epidemiological trends in nosocomial candidemia in intensive care. BMC Infect. Dis. 2006, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Hasan, F.; Xess, I.; Wang, X.; Jain, N.; Fries, B.C. Biofilm formation in clinical Candida isolates and its association with virulence. Microbes Infect. 2009, 11, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, V.; Barnes, A.J. Non-albicans Candida spp. Causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 2002, 50, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Krcmery, V. Torulopsis glabrata an emerging yeast pathogen in cancer patients. Int. J. Antimicrob. Agents 1999, 11, 1–6. [Google Scholar] [CrossRef]
- Tortorano, A.M.; Kibbler, C.; Peman, J.; Bernhardt, H.; Klingspor, L.; Grillot, R. Candidaemia in Europe: epidemiology and resistance. Int. J. Antimicrob. Agents 2006, 27, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Sardi, J.; Scorzoni, L.; Bernardi, T.; Fusco-Almeida, A.; Mendes Giannini, M. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013, 62, 10–24. [Google Scholar] [PubMed]
- Lim, C.S.-Y.; Rosli, R.; Seow, H.F.; Chong, P.P. Candida and invasive candidiasis: back to basics. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Abi-Said, D.; Anaissie, E.; Uzun, O.; Pinzcowski, H.; Vartivarian, S. The epidemiology of hematogeneous candidiasis caused by different Candida species. Clin. Infect. Dis. 1997, 24, 1122–1128. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Lozano-Chiu, M.; Rex, J. Declining rates of oropharyngeal candidiasis and carriage of Candida albicans associated with trends toward reduced rates of carriage of fluconazole-resistant C. albicans in human immunodeficiency virus-infected patients. Clin. Infect. Dis. 1998, 27, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Beck-Sague, C.; Jarvis, W. National nosocomial infectious surveillance system. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J. Infect. Dis. 1993, 167, 1247–1251. [Google Scholar] [CrossRef] [PubMed]
- Rees, J.; Pinner, R.; Hajjeh, R.; Brandt, M.; Reingold, A. The epidemiological features of invasive mycotic infections in the San Francisco Bay area, 1992–1993: results of population-based laboratory active surveillance. Clin. Infect. Dis. 1998, 7, 1138–1147. [Google Scholar] [CrossRef]
- Pfaller, M.; Jones, R.; Messer, S.; Edmond, M.; Wenzel, R. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn. Microbiol. Infect. Dis. 1998, 30, 121–129. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Jones, R.N.; Doern, G.V.; Sader, H.S.; Messer, S.A.; Houston, A.; Coffman, S.; Hollis, R.J. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997–1998. Antimicrob. Agents Chemother. 2000, 44, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Gumbo, T.; Isada, C.; Hall, G.; Karafa, M.; Gordon, S. Candida glabrata fungemia. Clinical features of 139 patients. Medicine 1999, 78, 220–227. [Google Scholar] [CrossRef]
- Pfaller, M.; Jones, R.; Doern, G.; Fluit, A.; Verhoef, J.; Sader, H.; Messer, S.; Houston, A.; Coffman, S.; Hollis, R. International surveillance of blood stream infections due to Candida species in the European SENTRY Program: species distribution and antifungal susceptibility including the investigational triazole and echinocandin agents. SENTRY Participant Group (Euro). Diagn. Microbiol. Infect. Dis. 1999, 35, 19–25. [Google Scholar] [CrossRef]
- Rodriguez-Tudela, J.; Cuenca-Estrella, M. Estudio multicéntrico sobre funguemias por levaduras en España (abril-junio de 1997). Grupo de trabajo para el estudio de las funguemias. Rev. Clin. Esp. 1999, 199, 356–361. [Google Scholar] [PubMed]
- Pfaller, M.; Diekema, D.; Jones, R.; Sader, H.; Fluit, A.; Hollis, R.; Messer, S.; Group, T.S.P. International surveillance of bloodstream infections due to Candida species: Frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 2001, 39, 3254–3259. [Google Scholar] [CrossRef] [PubMed]
- Wingard, J. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin. Infect. Dis. 1995, 20, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.; Pfaller, M.; Barry, A.; Nelson, P.; Webb, C. Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B as treatment of nonneutropenic patients with candidemia. Antimicrob. Agents Chemother. 1995, 39, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Abbas, J.; Bodey, G.P.; Hanna, H.A.; Mardani, M.; Girgawy, E.; Abi-Said, D.; Whimbey, E.; Hachem, R.; Raad, I. Candida krusei fungemia. An escalating serious infection in immunocompromised patients. Arch. Intern. Med. 2000, 160, 2659–2664. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, J.A.; Sanchez, V.; Dmuchowski, C.; Dembry, L.M.; Sobel, J.D.; Zervos, M.J. Nosocomial acquisition of Candida albicans: an epidemiologic study. J. Infect. Dis. 1993, 168, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, J.A.; Dembry, L.M.; Sanchez, V.; Vazquez, M.A.; Sobel, J.D.; Dmuchowski, C.; Zervos, M.J. Nosocomial Candida glabrata colonization: An epidemiologic study. J. Clin. Microbiol. 1998, 36, 421–426. [Google Scholar] [PubMed]
- Martins, M.; Henriques, M.; Ribeiro, A.P.; Fernandes, R.; Gonçalves, V.; Seabra, Á.; Azeredo, J.; Oliveira, R. Oral Candida carriage of patients attending a dental clinic in Braga, Portugal. Rev. Iberoam. Micol. 2010, 27, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masiá Canuto, M.; Gutiérrez Rodero, F.; Ortiz de la Tabla Ducasse, V.; Hernández Aguado, I.; Martín González, C.; Sánchez Sevillano, A.; Martín Hidalgo, A. Determinants for the development of oropharyngeal colonization or infection by fluconazole-resistant candida strains in HIV-Infected Patients. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Redding, S.W.; Kirkpatrick, W.R.; Coco, B.J.; Sadkowski, L.; Fothergill, A.W.; Rinaldi, M.G.; Eng, T.Y.; Patterson, T.F. Candida glabrata oropharyngeal candidiasis in patients receiving radiation treatment for head and neck cancer. J. Clin. Microbiol. 2002, 40, 1879–1881. [Google Scholar] [CrossRef] [PubMed]
- De Bernardis, F.; Chiani, P.; Ciccozzi, M.; Pellegrini, G.; Ceddia, T.; D’Offizzi, G.; Quinti, I.; Sullivan, P.A.; Cassone, A. Elevated aspartic proteinase secretion and experimental pathogenicity of Candida albicans isolates from oral cavities of subjects infected with human immunodeficiency virus. Infect. Immun. 1996, 64, 466–471. [Google Scholar] [PubMed]
- Fidel, P.L.; Cutright, J.L.; Tait, L.; Sobel, J.D. A murine model of Candida glabrata vaginitis. J. Infect. Dis. 1996, 173, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Negri, M.; Silva, S.; Henriques, M.; Oliveira, R. Insights into Candida tropicalis nosocomial infections and virulence factors. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1399–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, E.; Silva, S.; Rodrigues, C.F.; Alves, C.T.; Azeredo, J.; Henriques, M. Effects of fluconazole on Candida glabrata biofilms and its relationship with ABC transporter gene expression. Biofouling 2014, 30, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial Biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.; Costerton, J. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Silva, S.; Azeredo, J.; Henriques, M. Candida glabrata’s recurrent infections: biofilm formation during Amphotericin B treatment. Lett. Appl. Microbiol. 2016, 63, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Klotz, S.A.; Drutz, D.J.; Zajic, J.E. Factors Governing Adherence of Candida Species to Plastic Surfaces. Infect. Immun. 1985, 50, 97–101. [Google Scholar] [PubMed]
- Hazen, K.C.; Plotkin, B.J.; Klimas, D.M. Influence of growth conditions on cell surface hydrophobicity of Candida albicans and Candida glabrata. Infect. Immun. 1986, 54, 269–271. [Google Scholar] [PubMed]
- Kikutani, H.; Makino, S. The murine autoimmune diabetes model: NOD and related strains. Adv. Immunol. 1992, 51, 285–322. [Google Scholar] [PubMed]
- De Las Peñas, A.; Pan, S.J.; Castaño, I.; Alder, J.; Cregg, R.; Cormack, B.P. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003, 17, 2245–2258. [Google Scholar] [CrossRef] [PubMed]
- Castano, I.; Pan, S.; Zupancic, M.; Hennequin, C.; Dujon, B.; Cormack, B. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 2005, 55, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Sundstrom, P. Adhesion in Candida spp. Cell. Microbiol. 2002, 4, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Filler, S.G. Candida–host cell receptor–ligand interactions. Curr. Opin. Microbiol. 2006, 9, 333–339. [Google Scholar] [CrossRef] [PubMed]
- de Groot, P.W.J.; Kraneveld, E.A.; Yin, Q.Y.; Dekker, H.L.; Gross, U.; Crielaard, W.; de Koster, C.G.; Bader, O.; Klis, F.M.; Weig, M. The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 2008, 7, 1951–1964. [Google Scholar] [CrossRef] [PubMed]
- Kraneveld, E.A.; de Soet, J.J.; Deng, D.M.; Dekker, H.L.; de Koster, C.G.; Klis, F.M.; Crielaard, W.; de Groot, P.W.J. Identification and differential gene expression of adhesin-like wall proteins in Candida glabrata biofilms. Mycopathologia 2011, 172, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Adherence and biofilm formation of non-Candida albicans Candida species. Trends Microbiol. 2011, 19, 241–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcazar-Fuoli, L.; Mellado, E. Current status of antifungal resistance and its impact on clinical practice. Br. J. Haematol. 2014, 166. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, A.A.; Farley, M.M.; Harrison, L.H.; Stein, B.; Hollick, R.; Lockhart, S.R.; Magill, S.S.; Derado, G.; Park, B.J.; Chiller, T.M. Changes in incidence and antifungal drug resistance in candidemia: Results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clin. Infect. Dis. 2012, 55, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Shahi, P.; Moye-Rowley, W.S. Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim. Biophys. Acta 2009, 1794, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.; Thierry, A.; Coppée, J.-Y.; Gouyette, C.; Hennequin, C.; Sismeiro, O.; Talla, E.; Dujon, B.; Fairhead, C. Genomic polymorphism in the population of Candida glabrata: gene copy-number variation and chromosomal translocations. Fungal Genet. Biol. 2009, 46, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.; Bouchier, C.; Dujon, B.; Richard, G.-F. Megasatellites: a peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata. Nucleic Acids Res. 2008, 36, 5970–5982. [Google Scholar] [CrossRef] [PubMed]
- Barchiesi, F.; Falconi Di Francesco, L.; Arzeni, D.; Caselli, F.; Gallo, D.; Scalise, G. Electrophoretic Karyotyping and Triazole Susceptibility of Candida glabrata Clinical Isolates. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, C.S.; Merz, W.G. Electrophoretic karyotypes of Torulopsis glabrata. J. Clin. Microbiol. 1989, 27, 2165–2168. [Google Scholar] [PubMed]
- Klempp-Selb, B.; Rimek, D.; Kappe, R. Karyotyping of Candida albicans and Candida glabrata from patients with Candida sepsis. Mycoses 2000, 43, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Chen, Y.C.; Lo, H.J.; Chen, K.W.; Li, S.Y. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 2007, 45, 2452–2459. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Chae, M.J.; Song, J.W.; Jung, S.-I.; Cho, D.; Kee, S.J.; Kim, S.H.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 2007, 45, 2385–2391. [Google Scholar] [CrossRef] [PubMed]
- Magee, B.B.; Sanchez, M.D.; Saunders, D.; Harris, D.; Berriman, M.; Magee, P.T. Extensive chromosome rearrangements distinguish the karyotype of the hypovirulent species Candida dubliniensis from the virulent Candida albicans. Fungal Genet. Biol. 2008, 45, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Bader, O.; Schwarz, A.; Kraneveld, E.A.; Tangwattanachuleeporn, M.; Schmidt, P.; Jacobsen, M.D.; Gross, U.; De Groot, P.W.J.; Weig, M. Gross karyotypic and phenotypic alterations among different progenies of the Candida glabrata CBS138/ATCC2001 reference strain. PLoS ONE 2012, 7, e52218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rai, M.N.; Balusu, S.; Gorityala, N.; Dandu, L.; Kaur, R. Functional genomic analysis of Candida glabrata-macrophage interaction: role of chromatin remodeling in virulence. PLoS Pathog. 2012, 8, e1002863. [Google Scholar] [CrossRef] [PubMed]
- Jandric, Z.; Schuller, C. Stress response in Candida glabrata: pieces of a fragmented picture. Futur. Mirobiology 2011, 6, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Riera, M.; Mogensen, E.; d’Enfert, C.; Janbon, G. New regulators of biofilm development in Candida glabrata. Res. Microbiol. 2012, 163, 297–307. [Google Scholar] [CrossRef]
- Iraqui, I.; Garcia-Sanchez, S.; Aubert, S.; Dromer, F.; Ghigo, J.M.; d’Enfert, C.; Janbon, G. The YAK1P kinase controls expression of adhesins and biofilm formation in Candida glabrata in a SIR4P-dependent pathway. Mol. Microbiol. 2005, 55, 1259–1271. [Google Scholar] [CrossRef] [PubMed]
- Lemke, A.; Kiderlen, A.F.; Kayser, O. Amphotericin B. Appl. Microbiol. Biotechnol. 2005, 68, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Baginski, M.; Czub, J. Amphotericin B and its new derivatives-mode of action. Curr. Drug Metab. 2009, 10, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Botero, M.C.; Puentes-Herrera, M.; Cortés, J.A. Formas lipídicas de anfotericina. Rev. Chil. Infectol. 2014, 31, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Resistance to echinocandin- class antifungal drugs. Drug Resist. Updat. 2007, 10, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Debono, M.; Gordee, R.S. Antibiotics that inhibit fungal cell wall development. Annu. Rev. Microbiol. 1994, 48, 471–497. [Google Scholar] [CrossRef] [PubMed]
- Canuto, M.M.; Rodero, F.G. Antifungal drug resistance to azoles and polyenes. Lancet Infect. Dis. 2002, 2, 550–563. [Google Scholar] [CrossRef]
- Maenza, J.; Merz, W.; Romagnoli, M.; Keruly, J.; Moore, R.; Gallant, J. Infection due to fluconazole- resistant Candida in patients with AIDS: Prevalence and microbiology. Clin. Infect. Dis. 1997, 24, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Laguna, F.; Rodríguez-Tudela, J.L.; Martínez-Súarez, J.V.; Polo, R.; Valencia, E.; Díaz-Guerra, T.M.; Dronda, F.; Pulido, F.; Martínez-Suárez, J. Patterns of fluconazole susceptibility in isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis due to Candida albicans. Clin. Infect. Dis. 1997, 24, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Revankar, S.G.; Kirkpatrick, W.R.; McAtee, R.K.; Dib, O.P.; Fothergill, A.W.; Redding, S.W.; Rinaldi, M.G.; Patterson, T.F. Detection and significance of fluconazole resistance in oropharyngeal Candidiasis in human immunodeficiency virus-infected patients. J. Infect. Dis. 1996, 174, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Wingard, J.; Merz, W.; Rinaldi, M.; Jonhson, T.; Karp, J.; Saral, R. Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N. Engl. J. Med. 1991, 325, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, F.; Wall, P.; Cohen, J. Trends in antifungal use and antifungal isolates in a university hospital 1990–1992. J. Hosp. Infect. 1995, 31, 149–152. [Google Scholar] [CrossRef]
- Nguyen, M.L.H.; Peacock, J.E.; Morris, A.J.; Tanner, D.C.; Nguyen, M.L.H.; Snydman, D.R.; Wagener, M.M.; Rinaldi, M.G.; Yu, V.L. The changing face of candidemia: emergence of non- Candida albicans species and antifungal resistance. Am. J. Med. 1996, 100, 617–623. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Balzi, E.; Tulkens, P.M. Antibiotic Efflux Pumps. Biochem. Pharmacol. 2000, 60, 457–470. [Google Scholar] [CrossRef]
- Wilson, D.; Thewes, S.; Zakikhany, K.; Fradin, C.; Albrecht, A.; Almeida, R.; Brunke, S.; Grosse, K.; Martin, R.; Mayer, F.; et al. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res. 2009, 9, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Seider, K.; Almeida, R.; Heyken, A.; Fleck, C.; Brock, M.; Barz, D.; Rupp, S.; Hube, B. Candida glabrata tryptophan-based pigment production via the Ehrlich pathway. Mol. Microbiol. 2010, 76, 25–47. [Google Scholar] [CrossRef] [PubMed]
- Tscherner, M.; Schwarzmüller, T.; Kuchler, K. Pathogenesis and Antifungal Drug Resistance of the Human Fungal Pathogen Candida glabrata. Pharmaceuticals 2011, 4, 169–186. [Google Scholar] [CrossRef]
- Henry, K.; Nickels, J.; Edlind, T. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 2000, 44, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Stead, D.A.; Walker, J.; Holcombe, L.; Gibbs, S.R.S.; Yin, Z.; Selway, L.; Butler, G.; Brown, A.J.P.; Haynes, K. Impact of the transcriptional regulator, ACE2, on the Candida glabrata secretome. Proteomics 2010, 10, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, A.; Bignell, E.; Warn, P.; Jones, M.; Denning, D.; Muhlschlegel, F.; Rogers, T.; Haynes, K. C. glabrata STE12 is required for wild-type levels of virulence and nitrogen starvation induced filamentation. Mol. Microbiol. 2003, 50, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Kanafani, Z.A.; Perfect, J.R. Resistance to antifungal agents: mechanisms and clinical impact. Clin. Infect. Dis. 2008, 46, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sanguinetti, M.; Torelli, R.; Posteraro, B.; Sanglard, D. Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata. PLoS ONE 2011, 6, e17589. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sanguinetti, M.; De Bernardis, F.; Torelli, R.; Posteraro, B.; Vandeputte, P.; Sanglard, D. Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 2011, 55, 1852–1860. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.A.; Tsai, H.F.; Suffis, S.D.; Su, Q.; Myers, T.G.; Bennett, J.E. STB5 is a negative regulator of azole resistance in Candida glabrata. Antimicrob. Agents Chemother. 2013, 57, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Vermitsky, J.; Edlind, T.D. Azole Resistance in Candida glabrata: Coordinate upregulation of multidrug transporters and evidence for a PDR1-like transcription factor. Antimicrob. Agents Chemother. 2004, 48, 3773–3781. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.; Ischer, F.; Leibundgut-Landmann, S.; Sanglard, D. Gain-of-function mutations in PDR1, a regulator of antifungal drug resistance in Candida glabrata, control adherence to host cells. Infect. Immun. 2013, 81, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Schmidt, J.A.; Moye-Rowley, W.S. Regulation of the CGPDR1 transcription factor from the pathogen Candida glabrata. Eukaryot. Cell 2011, 10, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Farahyar, S.; Zaini, F.; Kordbacheh, P.; Rezaie, S.; Falahati, M.; Safara, M. Expression of efflux pumps and fatty acid activator one genes in azole resistant Candida glabrata isolated from immunocompromised patients. Acta Med. Iran. 2015, 54, 458–464. [Google Scholar]
- Kaur, R.; Castan, I.; Cormack, B.P. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: Roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 2004, 48, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Yamauchi, S.; Inamine, T.; Nagayoshi, Y.; Saijo, T.; Izumikawa, K.; Seki, M.; Kakeya, H.; Yamamoto, Y.; Yanagihara, K.; et al. Roles of calcineurin and Crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 2010, 54, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Ma, B.; Cormack, B.P. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc. Natl. Acad. Sci. USA. 2007, 104, 7628–7633. [Google Scholar] [CrossRef] [PubMed]
- Akins, R.A. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 2005, 43, 285–318. [Google Scholar] [CrossRef] [PubMed]
- Marichal, P.; Koymans, L.; Willemsens, S.; Bellens, D.; Verhasselt, P.; Luyten, W.; Borgers, M.; Ramaekers, F.C.; Odds, F.C.; Bossche, H.V. Contribution of mutations in the cytochrome P450 14-α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 1999, 145, 2701–2713. [Google Scholar] [CrossRef]
- Morio, F.; Pagniez, F.; Lacroix, C.; Miegeville, M.; Le Pape, P. Amino acid substitutions in the Candida albicans sterol D5,6-desaturase (Erg3p) confer azole resistance: characterization of two novel mutants with impaired virulence. J. Antimicrob. Chemother. 2012, 67, 2131–2138. [Google Scholar] [CrossRef] [PubMed]
- Szweda, P.; Gucwa, K.; Romanowska, E.; Dzierzanowska-Fangrat, K.; Naumiuk, L.; Brillowska-Dabrowska, A.; Wojciechowska-Koszko, I.; Milewski, S. Mechanisms of azole resistance among clinical isolates of Candida glabrata in Poland. J. Med. Microbiol. 2015, 64, 610–619. [Google Scholar] [CrossRef]
- Aramburu, J.; Rao, A.; Klee, C.B. Calcineurin: from structure to function. Curr. Top. Cell. Regul. 2000, 36, 237–295. [Google Scholar] [PubMed]
- Onyewu, C.; Blankenship, J.R.; Del Poeta, M.; Heitman, J. Ergosterol biosynthesis inhibitors become fungicidal when combined with calcineurin inhibitors against Candida albicans, Candida glabrata, and Candida krusei. Antimicrob. Agents Chemother. 2003, 47, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Cruz, M.C.; Goldstein, A.L.; Blankenship, J.R.; Del Poeta, M.; Davis, D.; Cardenas, M.E.; Perfect, J.R.; McCusker, J.H.; Heitman, J. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 2002, 21, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.H.; Miyazaki, T.; Tsai, H.F.; Bennett, J.E. The bZip transcription factor CGAP1P is involved in multidrug resistance and required for activation of multidrug transporter gene CgFLR1 in Candida glabrata. Gene 2007, 386, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Farahyar, S.; Zaini, F.; Kordbacheh, P.; Rezaie, S.; Safara, M.; Raoofian, R.; Heidari, M. Overexpression of Aldo-Keto-reductase in Azole-resistant clinical isolates of Candida glabrata determined by cDNA-AFLP. Daru J. Pharm. Sci. 2013, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Zarnowski, R.; Ross, K.M.; Sanchez, H.; Cain, M.T.; Hamaker, J.; Mitchell, A.P.; Andes, D.R. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 2012, 8, e1002848. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, J.; Rodriguez-Tudela, J.; Richard, C.; Alvarez, M.; Sanz, M.; Gaztelurrutia, L.; Ayats, J.; Martinez-Suarez, J. Deep infections caused by Scedosporium prolificans. A report on 16 cases in Spain and a review of the literature. Scedosporium prolificans Spanish Study Group. Medicine 1997, 76, 256–265. [Google Scholar] [CrossRef]
- Boutati, E.; Anaissie, E. Fusarium, a significant emerging pathogen in patients with hematologic malignancy: ten years’ experience at a cancer center and implications for management. Blood 1997, 90, 999–1008. [Google Scholar] [PubMed]
- Tritz, D.; Woods, G. Fatal disseminated infection with Aspergillus terreus in immunocompromised hosts. Clin. Infect. Dis. 1993, 16, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Rex, J.H.J.; Walsh, T.J.; Sobel, J.D.J.; Filler, S.G.; Pappas, P.G.; Dismukes, W.E.; Edwards, J.E. Practice guidelines for the treatment of candidiasis. Clin. Infect. Dis. 2000, 30, 662–678. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, P.; Tronchin, G.; Larcher, G.; Ernoult, E.; Bergès, T.; Chabasse, D.; Bouchara, J.-P. A nonsense mutation in the ERG6 gene leads to reduced susceptibility to polyenes in a clinical isolate of Candida glabrata. Antimicrob. Agents Chemother. 2008, 52, 3701–3709. [Google Scholar] [CrossRef] [Green Version]
- Vandeputte, P.; Tronchin, G.; Bergès, T.; Hennequin, C.; Chabasse, D.; Bouchara, J.P. Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrob. Agents Chemother. 2007, 51, 982–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Kwa, A.L.; Cheng, S.; Du, C.; Clancy, C.J. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 2012, 56, 4862–4869. [Google Scholar] [CrossRef] [PubMed]
- Beyda, N.D.; Lewis, R.E.; Garey, K.W. Echinocandin resistance in Candida species: mechanisms of reduced susceptibility and therapeutic approaches. Ann. Pharmacother. 2012, 46, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Gow, N.A.; Munro, C.A. Fungal echinocandin resistance. Fungal Genet. Biol. 2010, 47, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Barchiesi, F.; Barchiesi, F.; Spreghini, E.; Tomas- setti, S.; Della Vittoria, A.; Arzeni, D.; Manso, E.; Scalise, G. Effects of caspofungin against Candida guilliermondii and Candida parapsilosis. Antimicrob. Agents Chemother. 2006, 2719–2727. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Effron, G.; Katiyar, S.K.; Park, S.; Edlind, T.D.; Perlin, D.S. A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 2008, 52, 2305–2312. [Google Scholar] [CrossRef] [PubMed]
- Vai, M.; Orlandi, I.; Cavadini, P.; Alberghina, L.; Popolo, L. Candida albicans homologue of GGP1/GAS1 gene is functional in Saccharomyces cerevisiae and contains the determinants for glycosylphosphatidylinositol attachment. Yeast 1996, 12, 361–368. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Lee, S.; Park, S.; Cleary, J.D.; Perlin, D.S. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-β-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob. Agents Chemother. 2009, 53, 3690–3699. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jimenez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Thompson, G.R.; Wiederhold, N.P.; Vallor, A.C.; Villareal, N.C.; Lewis, J.S.; Patterson, T.F. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob. Agents Chemother. 2008, 52, 3783–3785. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.; Perlin, D.; Jensen, R.; Howard, S.; Goodwin, J.; Hopec, W. Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FSK resistance mutations. Antimicrob. Agents Chemother. 2012, 2435–2442. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.A.; White, T.C.; Perlin, D.S.; Selitrennikoff, C.P. Studies of the paradoxical effect of caspofungin at high drug concentrations. Diagn. Microbiol. Infect. Dis. 2005, 51, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Chamilos, G.; Lewis, R.E.; Albert, N.; Kontoyiannis, D.P. Paradoxical effect of echinocandins across candida species In vitro: Evidence for echinocandin-specific and candida species-related differences. Antimicrob. Agents Chemother. 2007, 51, 2257–2259. [Google Scholar] [CrossRef] [PubMed]
- Balashov, S.V.; Park, S.; Perlin, D.S. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 2006, 50, 2058–2063. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Rajendran, R.; Sherry, L.; Williams, C. Fungal biofilm resistance. Int. J. Microbiol. 2012, 2012, 528521. [Google Scholar] [CrossRef] [PubMed]
- Mathé, L.; Van Dijck, P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr. Genet. 2013, 59, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Mitchell, K.F.; Edward, J.A.; Andes, D.R. Mechanisms of Candida biofilm drug resistance. Futur. Microbiol. 2013, 8, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.; Singh-babak, S.; Hartooni, N.; Nobile, C. Biofilms and antifungal resistance. In Antifungals from Genomics to Resistance and the Development of Novel Agents; Coste, A., Vandeputte, P., Eds.; Caister Academic Press: Poole, UK, 2015; pp. 71–90. [Google Scholar]
- Anderson, J.B. Evolution of antifungal-drug resistance: Mechanisms and pathogen fitness. Nat. Rev. Microbiol. 2005, 3, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Jabra-Rizk, M.A.; Falkler, W.A.; Meiller, T.F. Fungal biofilms and drug resistance. Emerg. Infect. Dis. 2004, 10, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.; Lepak, A.; Marchillo, K.; Andes, D. Time course global gene expression analysis of an in vivo Candida biofilm. J. Infect. Dis. 2009, 200, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Vandewalle, K.; Wickes, B.; López-Ribot, J. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 2001, 18, 163–170. [Google Scholar] [PubMed]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.; Nett, J.; Oschel, P.; Albrecht, R.; Marchillo, K.; Pitula, A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 2004, 72, 6023–6031. [Google Scholar] [CrossRef] [PubMed]
- Mateus, C.; Crow, S.A.; Ahearn, D.G. Adherence of Candida albicans to silicone induces immediate enhanced tolerance to fluconazole. Antimicrob. Agents Chemother. 2004, 48, 3358–3366. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Bachmann, S.; Patterson, T.F.; Wickes, B.L.; López-Ribot, J.L. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 2002, 49, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Yeater, K.; Chandra, J.; Cheng, G.; Mukherjee, P.; Zhao, X.; Rodriguez- Zas, S.; Kwast, K.; Ghannoum, M.; Hoyer, L. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology 2007, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.H.; Yu, C.Y. Influence of incubation time, inoculum size, and glucose concentrations on spectrophotometric endpoint determinations for amphotericin B, fluconazole, and itraconazole. J. Clin. Microbiol. 1999, 37, 141–145. [Google Scholar] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing—Antifungal agents, Breakpoint tables for interpretation of MICs, Version 8.0, valid from 2015-11-16. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 1 December 2016).
- National Committee for Clinical Laboratory Standards. 1997. Available online: http://clsi.org/ (accessed on 1 December 2016).
- Perumal, P.; Mekala, S.; Chaffin, W.L. Role for cell density in antifungal drug resistance in Candida albicans biofilms. Antimicrob. Agents Chemother. 2007, 51, 2454–2463. [Google Scholar] [CrossRef] [PubMed]
- Hornby, J.; Jensen, E.; Lisec, A.; Tasto, J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Env. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed]
- Al-fattani, M.A.; Douglas, L.J. Penetration of candida biofilms by antifungal agents. Antimicrob. Agents Chemother. 2004, 48, 3291–3297. [Google Scholar] [CrossRef] [PubMed]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Lal, P.; Sharma, D.; Pruthi, P.; Pruthi, V. Exopolysaccharide analysis of biofilm-forming Candida albicans. J. Appl. Microbiol. 2010, 109, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Uppuluri, P.; Thomas, D.P.; Cleary, I.A.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia 2010, 169, 323–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, M.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 2012, 55, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Taff, H.T.; Cuevas, M.A.; Reinicke, E.L.; Sanchez, H.; Andes, D.R. Role of matrix β-1,3-glucan in antifungal resistance of non-albicans Candida biofilms. Antimicrob. Agents Chemother. 2013, 57, 1918–1920. [Google Scholar] [CrossRef] [PubMed]
- Vediyappan, G.; Rossignol, T.; D’Enfert, C. Interaction of Candida albicans biofilms with antifungals: Transcriptional response and binding of antifungals to β-glucans. Antimicrob. Agents Chemother. 2010. [Google Scholar] [CrossRef] [PubMed]
- Bandara, H.M.H.N.; Lam, O.L.T.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Bacterial lipopolysaccharides variably modulate in vitro biofilm formation of Candida species. J. Med. Microbiol. 2010, 59, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, S.C.; Smith, M.C.A.; Muston, P.; Holland, S.L.; Avery, S.V. Heterogeneous expression of the virulence-related adhesin EPA1 between individual cells and strains of the pathogen Candida glabrata. Eukaryot. Cell 2012, 11, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Kuhn, D.; Mukherjee, P.; Hoyer, L.; McCormick, T.; Ghannoum, M. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 2001, 183, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Baillie, G.S.; Douglas, L.J. Effect of growth rate on resistance of Candida albicans biofilms to antifungal agents. Antimicrob. Agents Chemother. 1998, 42, 1900–1905. [Google Scholar] [PubMed]
- Baillie, G.S.; Douglas, L.J. Iron-limited biofilms of Candida albicans and their susceptibility to amphotericin B. Antimicrob. Agents Chemother. 1998, 42, 2146–2149. [Google Scholar] [PubMed]
- Cannon, R.; Lamping, E.; Holmes, A. Candida albicans drug resistance—Another way to cope with stress. Microbiol 2007, 153, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, C.J.; Wang, Y.; Jin, L.; Abiko, Y.; Samaranayake, L.P. Proteomics of drug resistance in Candida glabrata biofilms. Proteomics 2010, 10, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [PubMed]
- Khot, P.D.; Suci, P.A.; Miller, R.L.; Nelson, R.D.; Tyler, B.J. A small subpopulation of blastospores in Candida albicans biofilms exhibit resistance to amphotericin B associated with differential regulation of ergosterol and β-1,6-glucan pathway genes. Antimicrob. Agents Chemother. 2006, 50, 3708–3716. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhaheri, R.S.; Douglas, L.J. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob. Agents Chemother. 2008, 52, 1884–1887. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.; Regenberg, B.; Gresham, D.; Folkesson, A. A common mechanism involving the TORC1 pathway can lead to amphotericin B-persistence in biofilm and planktonic Saccharomyces cerevisiae populations. Sci. Rep. 2016, 6, 21874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Farmakiotis, D.; Yang, D.; McCue, D.A.; Kantarjian, H.M.; Kontoyiannis, D.P.; Mathisen, M.S. The ever-evolving landscape of candidaemia in patients with acute leukaemia: non-susceptibility to caspofungin and multidrug resistance are associated with increased mortality. J. Antimicrob. Chemother. 2015. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Castanheira, M.; Lockhart, S.R.; Ahlquist, A.M.; Messer, S.A.; Jones, R.N. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J. Clin. Microbiol. 2012, 50, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, A.A.; Harrison, L.H.; Farley, M.M.; Hollick, R.; Stein, B.; Chiller, T.M.; Lockhart, S.R.; Park, B.J. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008-2013: Results from population-based surveillance. PLoS ONE 2015, 10, e0120452. [Google Scholar] [CrossRef] [PubMed]
- Razzaghi-Abyaneh, M.; Sadeghi, G.; Zeinali, E.; Alirezaee, M.; Shams-Ghahfarokhi, M.; Amani, A.; Mirahmadi, R.; Tolouei, R. Species distribution and antifungal susceptibility of Candida spp. isolated from superficial Candidiasis in outpatients in Iran. J. Mycol. Med. 2014, 24, e43–e50. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B.; Lass-Florl, C. Antifungal drug resistance among Candida species: Mechanisms and clinical impact. Mycoses 2015, 2, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Farmakiotis, D.; Tarrand, J.J.; Kontoyiannis, D.P. Drug-resistant Candida glabrata infection in cancer patients. Emerg. Infect. Dis. 2014, 20, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, J.; Motaghi, M.; Panahi, J.; Havasian, M.R.; Delpisheh, A.; Azizian, M.; Pakzad, I. Anti-fungal resistance in candida isolated from oral and diaper rash Candidiasis in neonates. Bioinformation 2014, 10, 667–670. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.C.; Tellería, O.; Cisterna, R. Sentinel surveillance of invasive candidiasis in Spain: Epidemiology and antifungal susceptibility. Diagn. Microbiol. Infect. Dis. 2015, 81, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Rhomberg, P.R.; Messer, S.A.; Jones, R.N.; Castanheira, M. Isavuconazole, micafungin, and 8 comparator antifungal agents’ susceptibility profiles for common and uncommon opportunistic fungi collected in 2013: temporal analysis of antifungal drug resistance using CLSI species-specific clinical breakpoints and proposed epidemiological cutoff values. Diagn. Microbiol. Infect. Dis. 2015, 82, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Schmalreck, A.F.; Willinger, B.; Haase, G.; Blum, G.; Lass-Flörl, C.; Fegeler, W.; Becker, K. Antifungal Susceptibility Testing-AFST Study Group Species and susceptibility distribution of 1062 clinical yeast isolates to azoles, echinocandins, flucytosine and amphotericin B from a multi-centre study. Mycoses 2012, 55, e124–e137. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.Y.; Yang, Y.P.; Zhang, Y.; Li, W.; Wang, J.D.; Huang, W.M.; Fan, Y.M. Molecular identification and antifungal susceptibility of 186 Candida isolates from vulvovaginal Candidiasis in Southern China. J. Med. Microbiol. 2015, 64, 390–393. [Google Scholar] [CrossRef] [PubMed]
- Zarei Mahmoudabadi, A.; Rezaei-Matehkolaei, A.; Ghanavati, F. The Susceptibility Patterns of Candida Species Isolated From Urine Samples to Posaconazole and Caspofungin. Jundishapur J. Microbiol. 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Spanu, T.; Fiori, B.; De Maio, F.; De Carolis, E.; Giaquinto, A.; Prete, V.; De Angelis, G.; Torelli, R.; D’Inzeo, T.; et al. Antifungal susceptibility profiles of bloodstream yeast isolates by sensititre yeastone over nine years at a large Italian teaching hospital. Antimicrob. Agents Chemother. 2015, 59, 3944–3955. [Google Scholar] [CrossRef] [PubMed]
- Czaika, V.; Nenoff, P.; Glöckner, A.; Becker, K.; Fegeler, W.; Schmalreck, A.F. Detection of azole susceptibility patterns in clinical yeast strains isolated from 1998 to 2008. New Microbiol. 2014, 37, 465–494. [Google Scholar] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Boyken, L.; Tendolkar, S.; Hollis, R.J.; Diekema, D.J. Geographic variation in the susceptibilities of invasive isolates of Candida glabrata to seven systemically active antifungal agents: A global assessment from the ARTEMIS Antifungal Surveillance Program conducted in 2001 and 2002. J. Clin. Microbiol. 2004, 42, 3142–3146. [Google Scholar] [CrossRef] [PubMed]
- Gopinathan, S.; Janagond, A.B.; Agatha, D.; P R, T. Detection of FUR1 Gene in 5-flucytosine resistant candida isolates in vaginal candidiasis patients. J. Clin. Diagn. Res. 2013, 7, 2452–2455. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, P.; Zareifar, S.; Badiee, P.; Alborzi, A.; Mokhtari, M.; Zomorodian, K.; Pakshir, K.; Jafarian, H. Yeast colonization and drug susceptibility pattern in the pediatric patients with neutropenia. Jundishapur J. Microbiol. 2014, 7, e11858. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, N.H.; Melake, N.A.; Somily, A.M.; Zakaria, A.S.; Baddour, M.M.; Mahmoud, A.Z. The effect of antifungal combination on transcripts of a subset of drug-resistance genes in clinical isolates of Candida species induced biofilms. Saudi Pharm. J. SPJ Off. Publ. Saudi Pharm. Soc. 2015, 23, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Andrade-Silva, L.; Ferreira-Paim, K.; Mora, D.J.; Da Silva, P.R.; Andrade, A.A.; Araujo, N.E.; Pedrosa, A.L.; Silva-Vergara, M.L. Susceptibility profile of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Uberaba, Minas Gerais, Brazil. Med. Mycol. 2013, 51, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Oxman, D.A.; Chow, J.K.; Frendl, G.; Hadley, S.; Hershkovitz, S.; Ireland, P.; McDermott, L.A.; Tsai, K.; Marty, F.M.; Kontoyiannis, D.P.; Golan, Y. Candidaemia associated with decreased in vitro fluconazole susceptibility: is Candida speciation predictive of the susceptibility pattern? J. Antimicrob. Chemother. 2010, 65, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Jones, R.N.; Castanheira, M. Antifungal susceptibilities of Candida, Cryptococcus neoformans and Aspergillus fumigatus from the Asia and Western Pacific region: Data from the SENTRY antifungal surveillance program (2010–2012). J. Antibiot. 2015, 68, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Chang, T.Y.; Liu, J.W.; Chen, F.J.; Chien, C.C.; Tang, Y.F.; Lu, C.H. Correlation of anti-fungal susceptibility with clinical outcomes in patients with cryptococcal meningitis. BMC Infect. Dis. 2012, 12, 361. [Google Scholar] [CrossRef] [PubMed]
- Matos, C.S.; de Souza Andrade, A.; Oliveira, N.S.; Barros, T.F. Microbiological characteristics of clinical isolates of Cryptococcus spp. in Bahia, Brazil: Molecular types and antifungal susceptibilities. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1647–1652. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, M.; Al-Sweih, N.A.; Ahmad, S.; Khan, Z.U. Antifungal susceptibility of clinical Candida parapsilosis isolates in Kuwait. Mycoses 2008, 51, 318–323. [Google Scholar] [CrossRef]
- Hegazi, M.; Abdelkader, A.; Zaki, M.; El-Deek, B. Characteristics and risk factors of candidemia in pediatric intensive care unit of a tertiary care children’s hospital in Egypt. J. Infect. Dev. Ctries. 2014, 8, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Zaoutis, T.E.; Foraker, E.; McGowan, K.L.; Mortensen, J.; Campos, J.; Walsh, T.J.; Klein, J.D. Antifungal susceptibility of Candida spp. isolated from pediatric patients: A survey of 4 children’s hospitals. Diagn. Microbiol. Infect. Dis. 2005, 52, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Krogh-Madsen, M.; Arendrup, M.C.; Heslet, L.; Knudsen, J.D. Amphotericin B and Ca- spofungin resistance in Candida glabrata isolates recovered from a critically ill patient. Clin. Infect. Dis. 2006, 42, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Rolley, N.; Kelly, D.E.; Kelly, S.L. Identification and characterization of four azole-resistant Erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 2010, 54, 4527–4533. [Google Scholar] [CrossRef] [PubMed]
- Eddouzi, J.; Parker, J.E.; Vale-Silva, L.A.; Coste, A.; Ischer, F.; Kelly, S.; Manai, M.; Sanglard, D. Molecular mechanisms of drug resistance in clinical Candida species isolated from Tunisian hospitals. Antimicrob. Agents Chemother. 2013, 3182–3193. [Google Scholar] [CrossRef] [PubMed]
- Forastiero, A.; Mesa-Arango, A.C.; Alastruey-Izquierdo, A.; Alcazar-Fuoli, L.; Bernal-Martinez, L.; Pelaez, T.; Lopez, J.F.; Grimalt, J.O.; Gomez- Lopez, A.; Cuesta, I.; Zaragoza, O.; Mellado, E. Candida tropicalis antifungal cross-resistance is related to different azole target (Erg11p) modifications. Antimicrob. Agents Chemother. 2013, 57, 4769–4781. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Pais, P.; Teixeira, M.C. Multidrug resistance in pathogenic yeasts: Emphasis on the role of ABC and MFS multidrug transporters. In The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs; Formatex: Badajoz, Spain, 2015; pp. 947–954. [Google Scholar]
- Sanglard, D.; Ischer, F.; Bille, J. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 2001, 45, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Torelli, R.; Posteraro, B.; Ferrari, S.; La Sorda, M.; Fadda, G.; Sanglard, D.; Sanguinetti, M. The ATP-binding cassette transporter-encoding gene CgSNQ2 is contributing to the CgPDR1-dependent azole resistance of Candida glabrata. Mol. Microbiol. 2008, 68, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Zhao, Y.; Perez, W.B.; Lockhart, S.R.; Sobel, J.D.; Farmakiotis, D.; Kontoyiannis, D.P.; Sanglard, D.; Taj-Aldeen, S.J.; Alexander, B.D.; et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016, 7, 11128. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Arthanari, H.; Yang, F.; Pan, S.J.; Fan, X.; Breger, J.; Frueh, D.P.; Gulshan, K.; Li, D.K.; Mylonakis, E.; et al. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 2008, 452, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Vought, B.W.; Satterlee, J.S.; Walker, A.K.; Sun, Z.-Y.J.; Watts, J.L.; DeBeaumont, R.; Mako Saito, R.; Hyberts, S.G.; Yang, S.; Macol, C.; et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 2006, 442, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.L.; Boeszoermenyi, A.; Vale-Silva, L.A.; Torelli, R.; Posteraro, B.; Sohn, Y.J.; Ji, F.; Gelev, V.; Sanglard, D.; Sanguinetti, M.; et al. Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 2016, 530, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Moye-Rowley, W.S. Multidrug resistance in fungi: Regulation of transporter-encoding gene expression. Front. Physiol. 2014, 5 APR, 143. [Google Scholar] [CrossRef] [PubMed]
- Orta-Zavalza, E.; Guerrero-Serrano, G.; Gutiérrez-Escobedo, G.; Cañas-Villamar, I.; Juárez-Cepeda, J.; Castaño, I.; De Las Peñas, A. Local silencing controls the oxidative stress response and the multidrug resistance in Candida glabrata. Mol. Microbiol. 2013, 88, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Pires, C.; Cabrito, T.R.; Renaudin, A.; Ohno, M.; Chibana, H.; Sá-Correia, I.; Teixeira, M.C. Candida glabrata drug:H+ antiporter CgQdr2 confers imidazole drug resistance, being activated by transcription factor CgPdr1. Antimicrob. Agents Chemother. 2013, 57, 3159–3167. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Nunes, J.; Henriques, A.; Mira, N.P.; Nakayama, H.; Chibana, H.; Teixeira, M.C. Candida glabrata drug:H+ antiporter CgTpo3 (ORF CAGL0I10384g): role in azole drug resistance and polyamine homeostasis. J. Antimicrob. Chemother. 2014, 69, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.; Henriques, A.; Pires, C.; Nunes, J.; Ohno, M.; Chibana, H.; Sá-Correia, I.; Teixeira, M.C. The dual role of Candida glabrata drug:H+ antiporter CgAqr1 (ORF CAGL0J09944g) in antifungal drug and acetic acid resistance. Front. Microbiol. 2013, 4, 170. [Google Scholar] [CrossRef] [PubMed]
- Hull, C.M.; Parker, J.E.; Bader, O.; Weig, M.; Gross, U.; Warrilow, A.G.S.; Kelly, D.E.; Kelly, S.L. Facultative sterol uptake in an ergosterol-deficient clinical isolate of Candida glabrata harboring a missense mutation in ERG11 and exhibiting cross-resistance to azoles and amphotericin B. Antimicrob. Agents Chemother. 2012, 56, 4223–4232. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.; Bergès, T.; Poupard, P.; Vauzelle-Moreau, C.; Renier, G.; Chabasse, D.; Bouchara, J.-P. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 2004, 48, 1788–1796. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Ischer, F.; Calabrese, D.; Posteraro, B.; Sanguinetti, M.; Fadda, G.; Rohde, B.; Bauser, C.; Bader, O.; Sanglard, D. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 2009, 5, e1000268. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, B.E.; Oltean, H.N.; Oliver, B.G.; Hoot, S.J.; Leyde, S.E.; Hedstrom, L.; White, T.C.; BE, M. Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 2010, 6, e1001126. [Google Scholar] [CrossRef] [PubMed]
- Miranda, I.M.; Silva, R.M.; Estevinho, M.; Pina-vaz, C. Environmental azole fungicide, prochloraz, can induce cross-resistance to medical triazoles in Candida glabrata. FEMS Yeast Res. 2014, 1–5. [Google Scholar]
- Helmerhorst, E.; Venuleo, C.; Sanglard, D.; Oppenheim, F. Roles of cellular respiration, CgCDR1, and CgCDR2 in Candida glabrata resistance to histatin. Antimicrob. Agents Chemother. 2006, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Edgerton, M.; Koshlukova, S. Salivary histatin 5 and its similarities to the other antimicrobial proteins in human saliva. Adv. Dent. Res. 2000, 14, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.; Hancock, R.; Devine, D.; Oppenheim, F. The antifungal mechanisms of antimicrobial proteins. In Mammalian Antimicrobial Proteins; Hancock, R., Devine, D., Eds.; Cambridge University Press: Cambridge, UK, 2004; pp. 245–277. [Google Scholar]
- Oppenheim, F. Salivary histidine-rich proteins. In Human Saliva: Clinical Chemistry and Microbiology; Tenovuo, J., Ed.; CRC Press: Boca Raton, FL, USA, 1989; pp. 151–160. [Google Scholar]
- Oppenheim, F.; Xu, T.; McMillian, F.; Levitz, S.; Diamond, R.; Offner, G.; Troxler, R. Histatins, a novel family of histidinerich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 1988, 263, 7472–7477. [Google Scholar] [PubMed]
- Tsai, H.; Bobek, L. Human salivary histatins: promising antifungal therapeutic agents. Crit. Rev. Oral Biol. Med. 1998, 9, 480–497. [Google Scholar] [CrossRef] [PubMed]
- van Urk, H.; Voll, W.; Scheffers, W.; van Dijken, J. Transient-state analysis of metabolic fluxes in Crabtree-positive and Crabtreenegative yeasts. Appl. Environ. Microbiol. 1990, 56, 281–287. [Google Scholar] [PubMed]
- Dalirsani, Z.; Adibpour, M.; Aghazadeh, M.; Amirchaghmaghi, M.; Falaki, F.; Mozafari, P.M.; Hamzei, F.M. In vitro comparison of inhibitory activity of 10 plant extracts against Candida albicans. Aust. J. Basic Appl. Sci. 2011, 5, 930–935. [Google Scholar]
- Junio, H.A.; Sy-Cordero, A.A.; Ettefagh, K.A.; Burns, J.T.; Micko, K.T.; Graf, T.N.; Richter, S.J.; Cannon, R.E.; Oberlies, N.H.; Cech, N.B. Synergy Directed Fractionation of Botanical Medicines: A Case Study with Goldenseal (Hydrastis canadensis). J. Nat. Prod. 2011, 74, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.; Fernandes Júnior, A. Biological properties of medicinal plants: A review of their antimicrobial activity. J. Venom. Anim. Toxins Incl. Trop. Dis. 2010, 16, 402–413. [Google Scholar] [CrossRef]
- Martins, N.; Barros, L.; Santos-Buelga, C.; Henriques, M.; Silva, S.; Ferreira, I.C.F.R. Evaluation of bioactive properties and phenolic compounds in different extracts prepared from Salvia officinalis L. Food Chem. 2015, 170, 378–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahanukar, S.A.; Kulkarni, R.A.; Rege, N.N. Pharmacology of medicinal plants and natural products. Indian J. Pharmacol. 2000, 32, S81–S118. [Google Scholar]
- Cowan, M. Plant Products as Antimicrobial Agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [PubMed]
- Sawamura, M. Aroma and functional properties of Japanese yuzu (Citrus junos Tanaka) essential oil. Aroma Res. 2000, 1, 14–19. [Google Scholar]
- Ormancey, X.; Sisalli, S.; Coutiere, P. Formulation of essential oils in functional parfumery. Parfum. Cosmet. Actual. 2001, 157, 30–40. [Google Scholar]
- Sharanappa, R.; Vidyasagar, G. Anti-Candida activity of medicinal plants. A review. Int. J. Pharm. Pharm. Sci. 2013, 5, 9–16. [Google Scholar]
- Djilani, A.; Dicko, A. The therapeutic benefits of essential oils. Nutrition, well-being and health. InTech 2012, 155–178. [Google Scholar]
- The Chemistry of Aromatherapeutic Oils, 3rd ed.; Bowles, J.E. (Ed.) Griffin Press: Salisbury, Australia, 2003.
- Gupta, V.; Mittal, P.; Bansal, P.; Khokra, S.L.; Kaushik, D. Pharmacological Potential of Matricaria recutita-A Review. Int. J. Pharm. Sci. Drug Res. 2010, 2, 12–16. [Google Scholar]
- Martín, Á.; Varona, S.; Navarrete, A.; Cocero, M.J. Encapsulation and co-precipitation processes with supercritical fluids: applications with essential oils. Open Chem. Eng. J. 2010, 4, 31–41. [Google Scholar] [CrossRef]
- Dorman, H.; Deans, S. Antimicrobial agents from plants: antibacterial activity of plant volatile oils. J. Appl. Microbiol. 2000, 88, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Aleksic, V.; Knezevic, P. Antimicrobial and antioxidative activity of extracts and essential oils of Myrtus communis L. Microbiol. Res. 2014, 169, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Aidi Wannes, W.; Mhamdi, B.; Sriti, J.; Ben Jemia, M.; Ouchikh, O.; Hamdaoui, G.; Kchouk, M.E.; Marzouk, B.; Wannes, A. Antioxidant activities of the essential oils and methanol extracts from myrtle (Myrtus communis var. Italica L.) leaf, stem and flower. Food Chem. Toxicol. 2010, 48, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Lapornik, B.; Prošek, M.; Wondra, A. Comparison of extracts prepared from plant by-products using different solvents and extraction time. J. Food Eng. 2005, 71, 214–222. [Google Scholar] [CrossRef]
- Pozzatti, P.; Scheid, L.; Spader, T.; Atayde, M.; Santurio, J.; Alves, S. In vitro activity of essential oils extracted from plants used as spices against fluconazole-resistant and fluconazole-susceptible Candida spp. Canadian. Can. J. Microbiol. 2008, 54, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Martins, N.; Ferreira, I.C.F.R.; Barros, L.; Carvalho, A.M.; Henriques, M.; Silva, S. Plants used in folk medicine: The potential of their hydromethanolic extracts against Candida species. Ind. Crops Prod. 2015, 66, 62–67. [Google Scholar] [CrossRef]
- Martins, N.; Ferreira, I.C.F.R.; Henriques, M.; Silva, S. In vitro anti-Candida activity of Glycyrrhiza glabra L. Ind. Crops Prod. 2016, 83, 81–85. [Google Scholar] [CrossRef]
- Prabhakar, K.; Kumar, L.S.; Rajendran, S.; Chandrasekaran, M.; Bhaskar, K.; Sajit Khan, A.K. Antifungal Activity of Plant Extracts against Candida Species from Oral Lesions. Indian J. Pharm. Sci. 2008, 70, 801–803. [Google Scholar] [CrossRef] [PubMed]
- Estevinho, L.; Afonso, S.; Feás, X. Antifungal effect of lavender honey against Candida albicans, Candida krusei and Cryptococcus neoformans. J. Food Sci. Technol. 2011, 48, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Estevinho, L.; Pereira, A.P.; Moreira, L.; Dias, L.G.; Pereira, E. Antioxidant and antimicrobial effects of phenolic compounds extracts of Northeast Portugal honey. Food Chem. Toxicol. 2008, 46, 3774–3779. [Google Scholar] [CrossRef] [PubMed]
- Bardy, J.; Nicholas, S.; Kathleen, M.; Alexander, M. A systematic review of honey uses and its potential value within oncology care. J. Clin. Nurs. 2008, 17, 2604–2623. [Google Scholar] [CrossRef] [PubMed]
- Lusby, E.; Coombes, A.; Wilkinson, J. Bactericidal Activity of Different Honeys against Pathogenic Bacteria. Arch. Med. Res. 2005, 36, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Molan, P. Why honey is effective as a medicine. Bee World 2001, 82, 22–40. [Google Scholar] [CrossRef]
- Eteraf-Oskouei, T.; Najafi, M. Traditional and Modern Uses of Natural Honey in Human Diseases: A Review. Iran. J. Basic Med. Sci. 2013, 16, 731–742. [Google Scholar] [PubMed]
- Theunissen, F.; Grobler, S.; Gedalia, I. The antifungal action of three South African honeys on Candida albicans. Apidologie 2001, 32, 371–379. [Google Scholar] [CrossRef]
- Irish, J.; Carter, D.; Shokohi, T.; Blair, S. Honey has an antifungal effect against Candida species. Med. Mycol. 2006, 44, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Küçük, M.; Kolaylı, S.; Karaoğlu, Ş.; Ulusoy, E.; Baltacı, C.; Candan, F. Biological activities and chemical composition of three honeys of different types from Anatolia. Food Chem. 2007, 100, 526–534. [Google Scholar] [CrossRef]
- Boukraa, L.; Benbarek, H.; Moussa, A. Synergistic action of starch and honey against Candida albicans in correlation with diastase number. Brazilian J. Microbiol. 2008, 39, 40–43. [Google Scholar] [CrossRef]
- Koc, A.N.; Silici, S.; Ercal, B.D.; Kasap, F.; Hörmet-Öz, H.T.; Mavus-Buldu, H. Antifungal Activity of Turkish Honey against Candida spp. and Trichosporon spp: An in vitro evaluation. Med. Mycol. 2009, 47, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Quadri, M.; Huraib, S. Manuka honey for central vein catheter exit site care. Semin. Dial. 1999, 12, 396–399. [Google Scholar] [CrossRef]
- Johnson, D.W.; van Eps, C.; Mudge, D.W.; Wiggins, K.J.; Armstrong, K.; Hawley, C.M.; Campbell, S.B.; Isbel, N.M.; Nimmo, G.R.; Gibbs, H. Randomized, controlled trial of topical exit-site application of honey (medihoney) versus mupirocin for the prevention of catheter-associated infections in hemodialysis patients. J. Am. Soc. Nephrol. 2005, 16, 1456–1462. [Google Scholar] [CrossRef] [PubMed]
- Freire, F.; Ferraresi, C.; Jorge, A.O.C.; Hamblin, M.R. Photodynamic therapy of oral Candida infection in a mouse model. J. Photochem. Photobiol. B Biol. 2016, 159, 161–168. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Mima, E.G.; Pavarina, A.C.; Dovigo, L.N.; Vergani, C.E.; de Souza Costa, C.A.; Kurachi, C.; Bagnato, V.S. Susceptibility of Candida albicans to photodynamic therapy in a murine model of oral candidosis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2010, 109, 392–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza, S.C.; Junqueira, J.C.; Balducci, I.; Koga-Ito, C.Y.; Munin, E.; Jorge, A.O.C. Photosensitization of different Candida species by low power laser light. J. Photochem. Photobiol. B Biol. 2006, 83, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.C.; Junqueira, J.C.; Rossoni, R.D.; Pereira, C.A.; Munin, E.; Jorge, A.O.C. Comparison of the photodynamic fungicidal efficacy of methylene blue, toluidine blue, malachite green and low-power laser irradiation alone against Candida albicans. Lasers Med. Sci. 2010, 25, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Nimrichter, L.; Nosanchuk, J.D.; Baltazar, L.M.; Ray, A.; Santos, D.A.; Cisalpino, P.S.; Friedman, A.J. Antimicrobial photodynamic therapy: an effective alternative approach to control fungal infections. Front. Microbiol. 2015, 6, 202. [Google Scholar]
- Giroldo, L.M.; Felipe, M.P.; de Oliveira, M.A.; Munin, E.; Alves, L.P.; Costa, M.S. Photodynamic antimicrobial chemotherapy (PACT) with methylene blue increases membrane permeability in Candida albicans. Lasers Med. Sci. 2009, 24, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Dovigo, L.N.; Pavarina, A.C.; de Oliveira Mima, E.G.; Giampaolo, E.T.; Vergani, C.E.; Bagnato, V.S. Fungicidal effect of photodynamic therapy against fluconazole-resistant Candida albicans and Candida glabrata. Mycoses 2011, 54, 123–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vartak, A.; Mutalik, V.; Parab, R.R.; Shanbhag, P.; Bhave, S.; Mishra, P.D.; Mahajan, G.B. Isolation of a new broad spectrum antifungal polyene from Streptomyces sp. MTCC 5680. Lett. Appl. Microbiol. 2014, 58, 591–596. [Google Scholar] [CrossRef] [PubMed]
- de Souza, T.B.; de Oliveira Brito, K.M.; Silva, N.C.; Rocha, R.P.; de Sousa, G.F.; Duarte, L.P.; Coelho, L.F.L.; Dias, A.L.T.; Veloso, M.P.; Carvalho, D.T.; et al. New eugenol glucoside-based derivative shows fungistatic and fungicidal activity against opportunistic Candida glabrata. Chem. Biol. Drug Des. 2015, 87, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Casanova, B.B.; Muniz, M.N.; Oliveira, T.; Oliveira, L.F.; Machado, M.M.; Fuentefria, A.M.; Gosmann, G.; Gnoatto, S.C.B. Synthesis and biological evaluation of some hydrazone derivatives as anti-inflammatory agents. Lett. Drug Des. Discov. 2012, 9, 310–315. [Google Scholar]
- Ferreira, B.D.S.; de Almeida, A.M.; Nascimento, T.C.; de Castro, P.P.; Silva, V.L.; Diniz, C.G.; Le Hyaric, M. Synthesis and biological evaluation of a new series of N-acyldiamines as potential antibacterial and antifungal agents. Bioorg. Med. Chem. Lett. 2014, 24, 4626–4629. [Google Scholar] [CrossRef] [PubMed]
- Abrão, P.H.O.; Pizi, R.B.; de Souza, T.B.; Silva, N.C.; Fregnan, A.M.; Silva, F.N.; Coelho, L.F.L.; Malaquias, L.C.C.; Dias, A.L.T.; Dias, D.F.; Veloso, M.P.; Carvalho, D.T. Synthesis and biological evaluation of new eugenol mannich bases as promising antifungal agents. Chem. Biol. Drug Des. 2015, 86, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.; Lee, M.-R.; Lynn, D.; Palecek, S. Antifungal Activity of 14-Helical β-peptides against planktonic cells and biofilms of Candida species. Pharmaceuticals 2015, 8, 483–503. [Google Scholar] [CrossRef] [PubMed]
- Silva-Dias, A.; Miranda, I.M.; Branco, J.; Cobrado, L.; Monteiro-Soares, M.; Pina-Vaz, C.; Rodrigues, A.G. In vitro antifungal activity and in vivo antibiofilm activity of cerium nitrate against Candida species. J. Antimicrob. Chemother. 2014, 70, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Joseph, M.R.P.; Al-Hakami, A.M.; Assiry, M.M.; Jamil, A.S.; Assiry, A. M.; Shaker, M.A.; Hamid, M.E. In vitro anti-yeast activity of chloramphenicol: A preliminary report. J. Med. Mycol. 2015, 25, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Ling, J.; Wu, C.D. Synergistic effects of tea catechin epigallocatechin gallate and antimycotics against oral Candida species. Arch. Oral Biol. 2015, 60, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Perera, J.; Weerasekera, M.; Kottegoda, N. Slow release anti-fungal skin formulations based on citric acid intercalated layered double hydroxides nanohybrids. Chem. Cent. J. 2015, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Pires, P.; Monteiro, D.R.; Negri, M.; Gorup, L.F.; Camargo, E.R.; Barbosa, D.B.; Oliveira, R.; Williams, D.W.; Henriques, M.; Azeredo, J. The effect of silver nanoparticles and nystatin on mixed biofilms of Candida glabrata and Candida albicans on acrylic. Med. Mycol. 2013, 51, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Casella, T.M.; Eparvier, V.; Mandavid, H.; Bendelac, A.; Odonne, G.; Dayan, L.; Duplais, C.; Espindola, L.S.; Stien, D. Antimicrobial and cytotoxic secondary metabolites from tropical leaf endophytes: Isolation of antibacterial agent pyrrocidine C from Lewia infectoria SNB-GTC2402. Phytochemistry 2013, 96, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Nirma, C.; Eparvier, V.; Stien, D. Antifungal Agents from Pseudallescheria boydii SNB-CN73 Isolated from a Nasutitermes sp. Termite. J. Nat. Prod. 2013, 76, 988–991. [Google Scholar] [CrossRef] [PubMed]
- Nirma, C.; Eparvier, V.; Stien, D. Antibacterial ilicicolinic acids c and d and ilicicolinal from neonectria discophora SNB-CN63 isolated from a termite nest. J. Nat. Prod. 2015, 78, 159–162. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, C.F.; Rodrigues, M.E.; Silva, S.; Henriques, M. Candida glabrata Biofilms: How Far Have We Come? J. Fungi 2017, 3, 11. https://doi.org/10.3390/jof3010011
Rodrigues CF, Rodrigues ME, Silva S, Henriques M. Candida glabrata Biofilms: How Far Have We Come? Journal of Fungi. 2017; 3(1):11. https://doi.org/10.3390/jof3010011
Chicago/Turabian StyleRodrigues, Célia F., Maria Elisa Rodrigues, Sónia Silva, and Mariana Henriques. 2017. "Candida glabrata Biofilms: How Far Have We Come?" Journal of Fungi 3, no. 1: 11. https://doi.org/10.3390/jof3010011







