Fungal Biofilms and Polymicrobial Diseases
Abstract
:1. Introduction
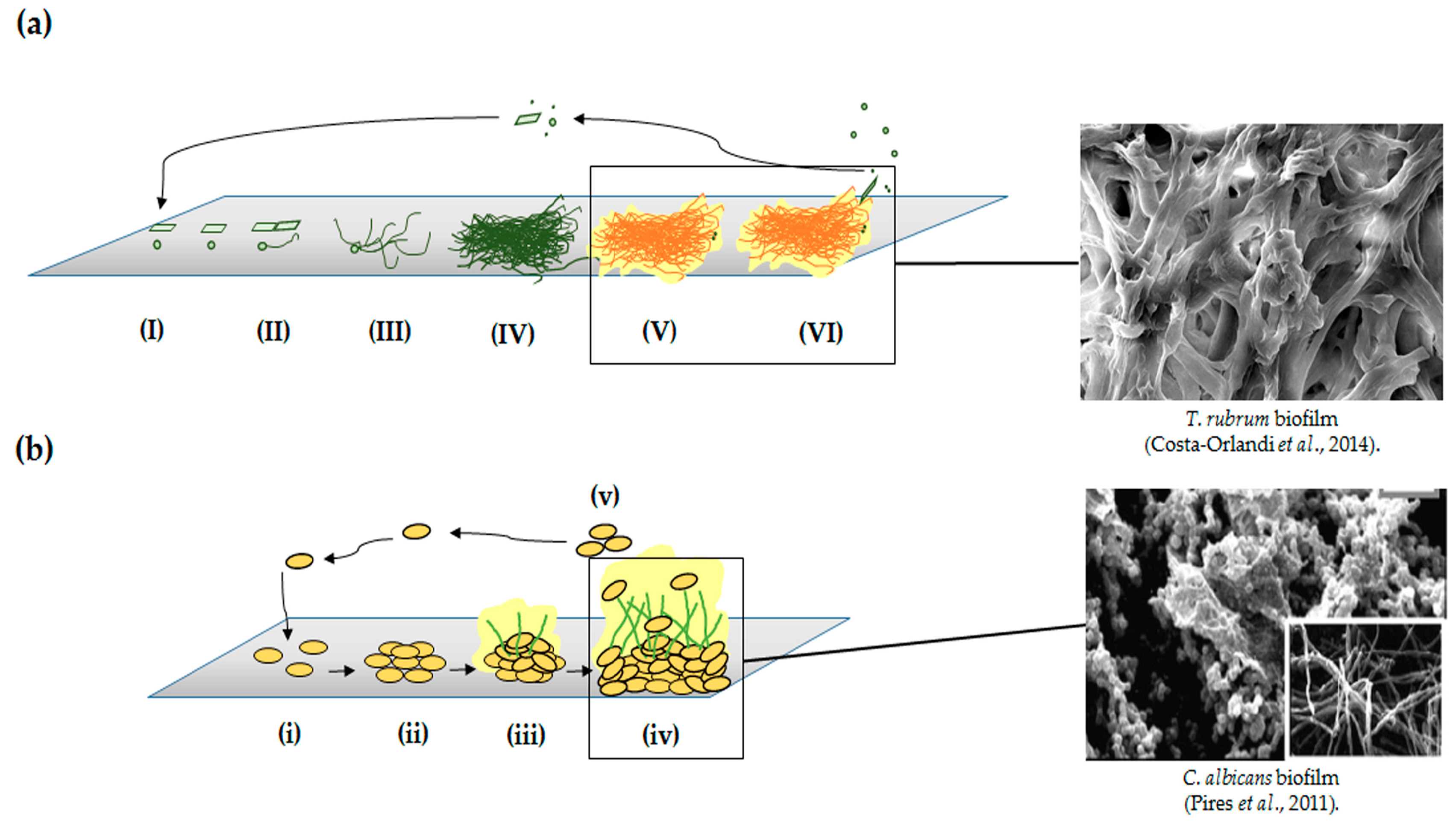
2. Yeasts and Filamentous Fungi Biofilms
3. Polymicrobial Biofilms
4. In Vitro Methods to Study Biofilms
4.1. Conventional Methods
4.2. High-Throughput “Omics” Technologies in Biofilms Research
5. In Vivo Models to Study Fungal Biofilms
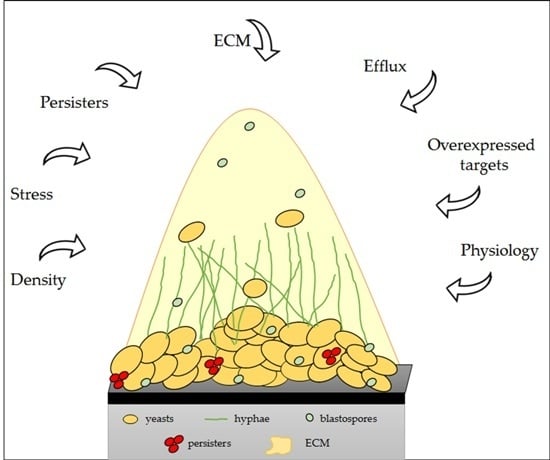
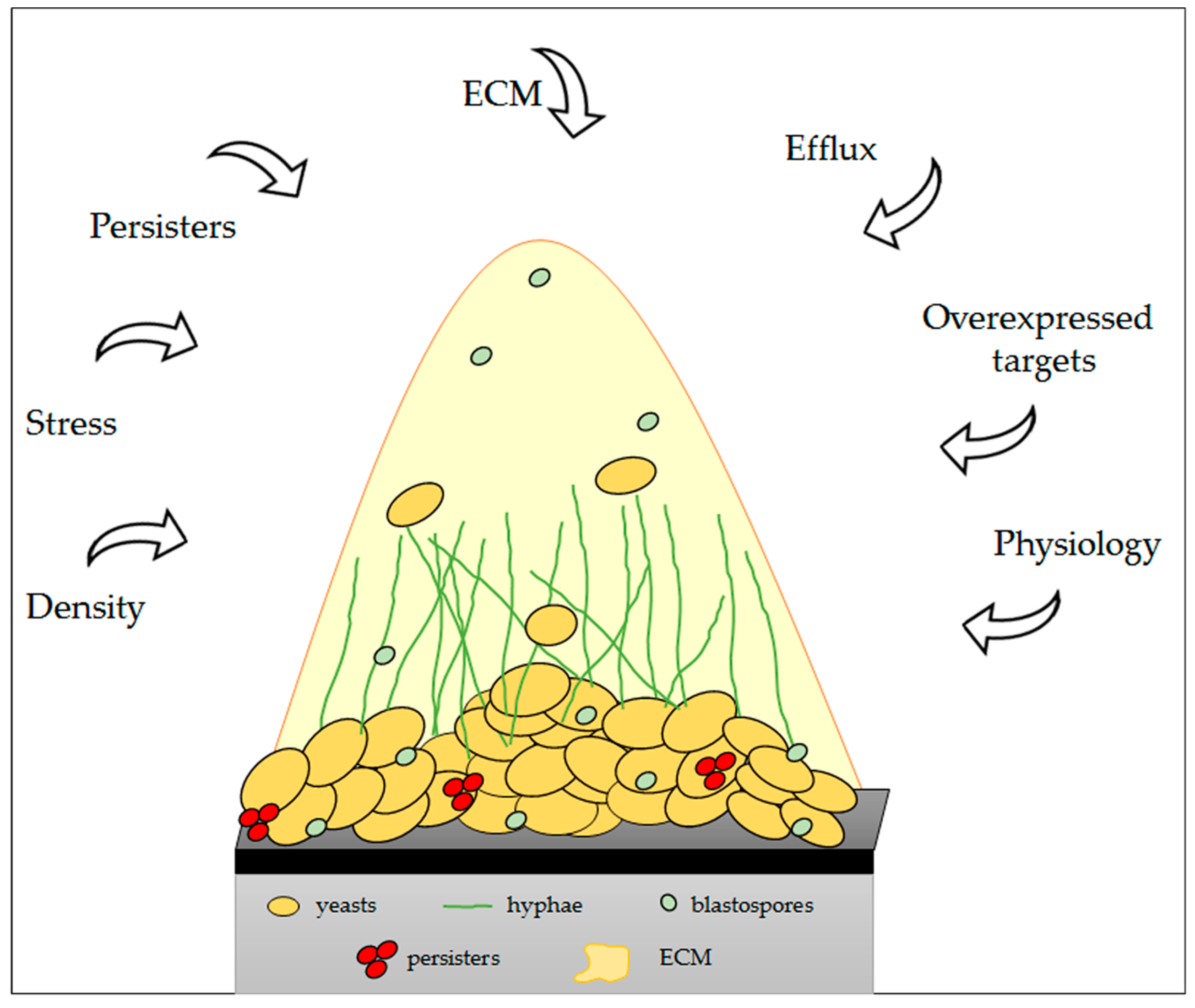
6. Physical and Molecular Resistance in Fungal Biofilms
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lopez-Ribot, J.L. Candida albicans biofilms: More than filamentation. Curr. Biol. 2005, 15, R453–R455. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Kuhn, D.M.; Mukherjee, P.K.; Hoyer, L.L.; McCormick, T.; Ghannoum, M.A. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J. Bacteriol. 2001, 183, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Fanning, S.; Mitchell, A.P. Fungal biofilms. PLoS Pathog. 2012, 8, e1002585. [Google Scholar] [CrossRef] [PubMed]
- Mowat, E.; Butcher, J.; Lang, S.; Williams, C.; Ramage, G. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J. Med. Microbiol. 2007, 56, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Pitangui, N.S.; Sardi, J.C.; Silva, J.F.; Benaducci, T.; Moraes da Silva, R.A.; Rodriguez-Arellanes, G.; Taylor, M.L.; Mendes-Giannini, M.J.; Fusco-Almeida, A.M. Adhesion of Histoplasma capsulatum to pneumocytes and biofilm formation on an abiotic surface. Biofouling 2012, 28, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.B.; Sardi, J.C.; Santos, C.T.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. In vitro characterization of Trichophyton rubrum and T. mentagrophytes biofilms. Biofouling 2014, 30, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Sardi Jde, C.; Pitangui Nde, S.; Voltan, A.R.; Braz, J.D.; Machado, M.P.; Fusco Almeida, A.M.; Mendes Giannini, M.J. In vitro Paracoccidioides brasiliensis biofilm and gene expression of adhesins and hydrolytic enzymes. Virulence 2015, 6, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Sardi Jde, C.; Pitangui Nde, S.; Rodriguez-Arellanes, G.; Taylor, M.L.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. Highlights in pathogenic fungal biofilms. Rev. Iberoam. Micol. 2014, 31, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.W.; Marques, L.L.; Howard, R.J.; Olson, M.E. Can filamentous fungi form biofilms? Trends Microbiol. 2009, 17, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Fries, B.C. Fungal biofilms: Relevance in the setting of human disease. Curr. Fungal Infect. Rep. 2010, 4, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, J.R.; Mitchell, A.P. How to build a biofilm: A fungal perspective. Curr. Opin. Microbiol. 2006, 9, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Wosten, H.A. Hydrophobins: Multipurpose proteins. Annu. Rev. Microbiol. 2001, 55, 625–646. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.; Andes, D. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr. Opin. Microbiol. 2006, 9, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.H.; Montanari, L.B.; Martins, C.H.; Zaia, J.E.; Almeida, A.M.; Matsumoto, M.T.; Mendes-Giannini, M.J. Anticandidal efficacy of cinnamon oil against planktonic and biofilm cultures of Candida parapsilosis and Candida orthopsilosis. Mycopathologia 2011, 172, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Weitzman, I.; Summerbell, R.C. The dermatophytes. Clin. Microbiol. Rev. 1995, 8, 240–259. [Google Scholar] [CrossRef]
- Costa-Orlandi, C.B.; Magalhães, G.M.; Oliveira, M.B.; Taylor, E.L.; Marques, C.R.; de Resende-Stoianoff, M.A. Prevalence of dermatomycosis in a Brazilian tertiary care hospital. Mycopathologia 2012, 174, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, C.N.; Burkhart, C.G.; Gupta, A.K. Dermatophytoma: Recalcitrance to treatment because of existence of fungal biofilm. J. Am. Acad. Dermatol. 2002, 47, 629–631. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses 2012, 55, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; George, T.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Antifungal susceptibility of Candida biofilms: Unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 2002, 46, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Hawser, S.P.; Douglas, L.J. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect. Immun. 1994, 62, 915–921. [Google Scholar] [PubMed]
- Marcos-Zambrano, L.J.; Gomez-Perosanz, M.; Escribano, P.; Zaragoza, O.; Bouza, E.; Guinea, J. Biofilm production and antibiofilm activity of echinocandins and liposomal amphotericin B in echinocandin-resistant yeast species. Antimicrob. Agents Chemother. 2016, 60, 3579–3586. [Google Scholar] [CrossRef] [PubMed]
- Martins, C.H.; Pires, R.H.; Cunha, A.O.; Pereira, C.A.; Singulani, J.L.; Abrão, F.; Moraes, T.; Mendes-Giannini, M.J. Candida/candida biofilms. First description of dual-species Candida albicans/C. rugosa biofilm. Fungal Biol. 2016, 120, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, H.; Shang, Q.; Jiang, Y.; Cao, Y.; Chai, Y. Time course analysis of Candida albicans metabolitesduring biofilm development. J. Proteome Res. 2013, 12, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.H.; Cataldi, T.R.; Franceschini, L.M.; Labate, M.V.; Fusco-Almeida, A.M.; Labate, C.A.; Palma, M.S.; Soares Mendes-Giannini, M.J. Metabolic profiles of planktonic and biofilm cells of Candida orthopsilosis. Future Microbiol. 2016, 11, 1299–1313. [Google Scholar] [CrossRef] [PubMed]
- Ricicová, M.; Kucharíková, S.; Tournu, H.; Hendrix, J.; Bujdáková, H.; Van Eldere, J.; Lagrou, K.; van Dijck, P. Candida albicans biofilm formation in a new in vivo rat model. Microbiology 2010, 156, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Kucharikova, S.; Vande Velde, G.; Himmelreich, U.; van Dijck, P. Candida albicans biofilm development on medically-relevant foreign bodies in a mouse subcutaneous model followed by bioluminescence imaging. J. Vis. Exp. 2015, 52239. [Google Scholar] [CrossRef] [PubMed]
- Henriques, M.; Azeredo, J.; Oliveira, R. Candida albicans and Candida dubliniensis: Comparison of biofilm formation in terms of biomass and activity. Br. J. Biomed. Sci. 2006, 63, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.H.; Santos, J.M.; Zaia, J.E.; Martins, C.H.; Mendes-Giannini, M.J. Candida parapsilosis complex water isolates from a haemodialysis unit: Biofilm production and in vitro evaluation of the use of clinical antifungals. Memórias Inst. Oswaldo Cruz 2011, 106, 646–654. [Google Scholar] [CrossRef]
- Bonhomme, J.; d’Enfert, C. Candida albicans biofilms: Building a heterogeneous, drug-tolerant environment. Curr. Opin. Microbiol. 2013, 16, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. In Nature Reviews. Microbiology; Nature Publishing Group: London, UK, 2011; Volume 9, pp. 109–118. [Google Scholar]
- Paramonova, E.; Krom, B.P.; van der Mei, H.C.; Busscher, H.J.; Sharma, P.K. Hyphal content determines the compression strength of Candida albicans biofilms. Microbiology 2009, 155, 1997–2003. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Martins, A.; Oliveira, R.; Williams, D.; Azeredo, J. Biofilms of non-Candida albicans Candida species: Quantification, structure and matrix composition. Med. Mycol. 2009, 47, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ramirez, A.I.; Ramirez-Granillo, A.; Medina-Canales, M.G.; Rodriguez-Tovar, A.V.; Martinez-Rivera, M.A. Analysis and description of the stages of Aspergillus fumigatus biofilm formation using scanning electron microscopy. BMC Microbiol. 2016, 16, 243. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.M.; Seidler, M.; Beauvais, A. Aspergillus fumigatus biofilms in the clinical setting. Med. Mycol. 2011, 49, S96–S100. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Singh, S. Biofilm formation by Aspergillus fumigatus. Med. Mycol. 2014, 52, 2–9. [Google Scholar] [PubMed]
- Ramage, G.; Rajendran, R.; Gutierrez-Correa, M.; Jones, B.; Williams, C. Aspergillus biofilms: Clinical and industrial significance. FEMS Microbiol. Lett. 2011, 324, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Rajendran, R.; Ramage, G. Aspergillus biofilms in human disease. Adv. Exp. Med. Biol. 2016, 931, 1–11. [Google Scholar] [PubMed]
- Seidler, M.J.; Salvenmoser, S.; Muller, F.M. Aspergillus fumigatus forms biofilms with reduced antifungal drug susceptibility on bronchial epithelial cells. Antimicrob. Agents Chemother. 2008, 52, 4130–4136. [Google Scholar] [CrossRef] [PubMed]
- Villena, G.K.; Fujikawa, T.; Tsuyumu, S.; Gutierrez-Correa, M. Structural analysis of biofilms and pellets of Aspergillus niger by confocal laser scanning microscopy and cryo scanning electron microscopy. Bioresour. Technol. 2010, 101, 1920–1926. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Schmidt, C.; Guadagnini, S.; Roux, P.; Perret, E.; Henry, C.; Paris, S.; Mallet, A.; Prevost, M.C.; Latge, J.P. An extracellular matrix glues together the aerial-grown hyphae of Aspergillus fumigatus. Cell. Microbiol. 2007, 9, 1588–1600. [Google Scholar] [CrossRef] [PubMed]
- Ajesh, K.; Sreejith, K. Cryptococcus laurentii biofilms: Structure, development and antifungal drug resistance. Mycopathologia 2012, 174, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Casadevall, A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 2006, 50, 1021–1033. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K.; Repp, K.K.; Hazen, K.C. Temperature affects the susceptibility of Cryptococcus neoformans biofilms to antifungal agents. Med. Mycol. 2010, 48, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Mihu, M.R.; Han, G.; Frases, S.; Cordero, R.J.; Casadevall, A.; Friedman, A.J.; Friedman, J.M.; Nosanchuk, J.D. The use of chitosan to damage Cryptococcus neoformans biofilms. Biomaterials 2010, 31, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Bumroongthai, K.; Chetanachan, P.; Niyomtham, W.; Yurayart, C.; Prapasarakul, N. Biofilm production and antifungal susceptibility of co-cultured Malassezia pachydermatis and Candida parapsilosis isolated from canine seborrheic dermatitis. Med. Mycol. 2016, 54, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Iturrieta-Gonzalez, I.A.; Padovan, A.C.; Bizerra, F.C.; Hahn, R.C.; Colombo, A.L. Multiple species of trichosporon produce biofilms highly resistant to triazoles and amphotericin B. PLoS ONE 2014, 9, e109553. [Google Scholar] [CrossRef] [PubMed]
- Imamura, Y.; Chandra, J.; Mukherjee, P.K.; Lattif, A.A.; Szczotka-Flynn, L.B.; Pearlman, E.; Lass, J.H.; O’Donnell, K.; Ghannoum, M.A. Fusarium and Candida albicans biofilms on soft contact lenses: Model development, influence of lens type, and susceptibility to lens care solutions. Antimicrob. Agents Chemother. 2008, 52, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Chandra, J.; Yu, C.; Sun, Y.; Pearlman, E.; Ghannoum, M.A. Characterization of fusarium keratitis outbreak isolates: Contribution of biofilms to antimicrobial resistance and pathogenesis. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4450–4457. [Google Scholar] [CrossRef] [PubMed]
- Peiqian, L.; Xiaoming, P.; Huifang, S.; Jingxin, Z.; Ning, H.; Birun, L. Biofilm formation by Fusarium oxysporum f. Sp. Cucumerinum and susceptibility to environmental stress. FEMS Microbiol. Lett. 2013, 350, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Mello, T.P.; Aor, A.C.; Goncalves, D.S.; Seabra, S.H.; Branquinha, M.H.; Santos, A.L. Assessment of biofilm formation by Scedosporium apiospermum, S. aurantiacum, S. minutisporum and Lomentospora prolificans. Biofouling 2016, 32, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.E.; Cook, G.; Costerton, J.W. Biofilm on ventriculo-peritoneal shunt tubing as a cause of treatment failure in coccidioidal meningitis. Emerg. Infect. Dis. 2002, 8, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Galván, E.M.; Mateyca, C.; Ielpi, L. Role of interspecies interactions in dual-species biofilms developed in vitro by uropathogens isolated from polymicrobial urinary catheter-associated bacteriuria. Biofouling 2016, 32, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Stacy, A.; McNally, L.; Darch, S.E.; Brown, S.P.; Whiteley, M. The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 2016, 14, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Willems, H.M.; Xu, Z.; Peters, B.M. Polymicrobial biofilm studies: From basic science to biofilm control. Curr. Oral Health Rep. 2016, 3, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The polymicrobial nature of biofilm infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.L.; Connell, J.L.; Stacy, A.; Turner, K.H.; Whiteley, M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 2014, 52, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Rendueles, O.; Ghigo, J.M. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Ren, D.; Bjarnsholt, T.; Sørensen, S.J. Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Short, F.L.; Murdoch, S.L.; Ryan, R.P. Polybacterial human disease: The ills of social networking. Trends Microbiol. 2014, 22, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Gabrilska, R.A.; Rumbaugh, K.P. Biofilm models of polymicrobial infection. Future Microbiol. 2015, 10, 1997–2015. [Google Scholar] [CrossRef] [PubMed]
- Jemielita, M.; Taormina, M.J.; Burns, A.R.; Hampton, J.S.; Rolig, A.S.; Guillemin, K.; Parthasarathy, R. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Monier, J.M.; Lindow, S.E. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. USA 2003, 100, 15977–15982. [Google Scholar] [CrossRef] [PubMed]
- Connell, J.L.; Wessel, A.K.; Parsek, M.R.; Ellington, A.D.; Whiteley, M.; Shear, J.B. Probing prokaryotic social behaviors with bacterial “Lobster traps”. mBio 2010, 1, e00202–e00210. [Google Scholar] [CrossRef] [PubMed]
- Blanchette-Cain, K.; Hinojosa, C.A.; Akula Suresh Babu, R.; Lizcano, A.; Gonzalez-Juarbe, N.; Munoz-Almagro, C.; Sanchez, C.J.; Bergman, M.A.; Orihuela, C.J. Streptococcus pneumoniae biofilm formation is strain dependent, multifactorial, and associated with reduced invasiveness and immunoreactivity during colonization. mBio 2013, 4, e00745-13. [Google Scholar] [CrossRef] [PubMed]
- Guggenberger, C.; Wolz, C.; Morrissey, J.A.; Heesemann, J. Two distinct coagulase-dependent barriers protect Staphylococcus aureus from neutrophils in a three dimensional in vitro infection model. PLoS Pathog. 2012, 8, e1002434. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.N.; Alhede, M.; Jensen, P.; Moser, C.; Scheike, T.; Jacobsen, C.S.; Seier Poulsen, S.; Eickhardt-Sørensen, S.R.; Trøstrup, H.; Christoffersen, L.; et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 2014, 82, 4477–4486. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Mohammadi, S.; Isberg, R.R. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe 2015, 17, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Weimer, K.E.; Juneau, R.A.; Murrah, K.A.; Pang, B.; Armbruster, C.E.; Richardson, S.H.; Swords, W.E. Divergent mechanisms for passive pneumococcal resistance to β-lactam antibiotics in the presence of haemophilus influenzae. J. Infect. Dis. 2011, 203, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Sonnenburg, J.L. Eating for two: How metabolism establishes interspecies interactions in the gut. Cell Host Microbe 2011, 10, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, M.S.; Mostow, E.; Mukherjee, P.; Hu, F.Z.; Melton-Kreft, R.; Ehrlich, G.D.; Dowd, S.E.; Ghannoum, M.A. Characterization of bacterial communities in venous insufficiency wounds by use of conventional culture and molecular diagnostic methods. J. Clin. Microbiol. 2011, 49, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, G.D.; Hu, F.Z.; Shen, K.; Stoodley, P.; Post, J.C. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin. Orthop. Relat. Res. 2005, 20–24. [Google Scholar] [CrossRef]
- Kolenbrander, P.E.; Palmer, R.J.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Dixon, E.F.; Hall, R.A. Noisy neighbourhoods: Quorum sensing in fungal-polymicrobial infections. Cell. Microbiol. 2015, 17, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, L.E.; Millhouse, E.; Sherry, L.; Kean, R.; Malcolm, J.; Nile, C.J.; Ramage, G. Polymicrobial Candida biofilms: Friends and foe in the oral cavity. FEMS Yeast Res. 2015, 15, fov077. [Google Scholar] [CrossRef] [PubMed]
- Jack, A.A.; Daniels, D.E.; Jepson, M.A.; Vickerman, M.M.; Lamont, R.J.; Jenkinson, H.F.; Nobbs, A.H. Streptococcus gordonii comcde (competence) operon modulates biofilm formation with Candida albicans. Microbiology 2015, 161, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Rautemaa, R.; Ramage, G. Oral candidosis—Clinical challenges of a biofilm disease. Crit. Rev. Microbiol. 2011, 37, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Dongari-Bagtzoglou, A.; Kashleva, H.; Dwivedi, P.; Diaz, P.; Vasilakos, J. Characterization of mucosal Candida albicans biofilms. PLoS ONE 2009, 4, e7967. [Google Scholar] [CrossRef] [PubMed]
- Dutton, L.C.; Jenkinson, H.F.; Lamont, R.J.; Nobbs, A.H. Role of Candida albicans secreted aspartyl protease Sap9 in interkingdom biofilm formation. Pathog. Dis. 2016, 74, ftw005. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Vickerman, M.M.; Jenkinson, H.F. Heterologous expression of Candida albicans cell wall-associated adhesins in saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryot. Cell 2010, 9, 1622–1634. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.J.; Nobbs, A.H.; Vickerman, M.M.; Barbour, M.E.; Jenkinson, H.F. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 2010, 78, 4644–4652. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Sobue, T.; Thompson, A.; Xie, Z.; Poon, K.; Ricker, A.; Cervantes, J.; Diaz, P.I.; Dongari-Bagtzoglou, A. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 2014, 16, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.R.; van der Wielen, P.; Cannon, R.D.; Ruske, D.; Dawes, P. Candida albicans binds to saliva proteins selectively adsorbed to silicone. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2006, 102, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Zago, C.E.; Silva, S.; Sanitá, P.V.; Barbugli, P.A.; Dias, C.M.; Lordello, V.B.; Vergani, C.E. Dynamics of biofilm formation and the interaction between Candida albicans and methicillin-susceptible (MSSA) and -resistant Staphylococcus aureus (MRSA). PLoS ONE 2015, 10, e0123206. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Jabra-Rizk, M.A.; Scheper, M.A.; Leid, J.G.; Costerton, J.W.; Shirtliff, M.E. Microbial interactions and differential protein expression in Staphylococcus aureus-Candida albicans dual-species biofilms. FEMS Immunol. Med. Microbiol. 2010, 59, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Ishihara, K.; Okuda, K. Prevalence of potential respiratory pathogens in the mouths of elderly patients and effects of professional oral care. Arch. Gerontol. Geriatr. 2001, 32, 45–55. [Google Scholar] [CrossRef]
- Baena-Monroy, T.; Moreno-Maldonado, V.; Franco-Martínez, F.; Aldape-Barrios, B.; Quindós, G.; Sánchez-Vargas, L.O. Candida albicans, Staphylococcus aureus and streptococcus mutans colonization in patients wearing dental prosthesis. Med. Oral Patol. Oral Cir. Bucal 2005, 10, E27–E39. [Google Scholar] [PubMed]
- Miyake, Y.; Iwai, T.; Sugai, M.; Miura, K.; Suginaka, H.; Nagasaka, N. Incidence and characterization of Staphylococcus aureus from the tongues of children. J. Dent. Res. 1991, 70, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Robertson, D.; Tang, M.K.; Jackson, M.S.; MacKenzie, D.; Bagg, J. Staphylococcus aureus in the oral cavity: A three-year retrospective analysis of clinical laboratory data. Br. Dent. J. 2003, 195, 701–703. [Google Scholar] [CrossRef] [PubMed]
- Ohara-Nemoto, Y.; Haraga, H.; Kimura, S.; Nemoto, T.K. Occurrence of staphylococci in the oral cavities of healthy adults and nasal oral trafficking of the bacteria. J. Med. Microbiol. 2008, 57, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Coco, B.J.; Bagg, J.; Cross, L.J.; Jose, A.; Cross, J.; Ramage, G. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol. Immunol. 2008, 23, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Henriques, M.; Hayes, A.; Oliveira, R.; Azeredo, J.; Williams, D.W. Candida glabrata and Candida albicans co-infection of an in vitro oral epithelium. J. Oral Pathol. Med. 2011, 40, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.T.; Wei, X.Q.; Silva, S.; Azeredo, J.; Henriques, M.; Williams, D.W. Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J. Infect. 2014, 69, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.T.; Tan, H.W.; Na, S.L.; Lim, S.L. Phenotypic and genotypic characterization of two closely related subgroups of Candida rugosa in clinical specimens. J. Med. Microbiol. 2011, 60, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Colombo, A.L.; Melo, A.S.; Crespo Rosas, R.F.; Salomão, R.; Briones, M.; Hollis, R.J.; Messer, S.A.; Pfaller, M.A. Outbreak of Candida rugosa candidemia: An emerging pathogen that may be refractory to amphotericin B therapy. Diagn. Microbiol. Infect. Dis. 2003, 46, 253–257. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A.; Pfaller, M.A.; Bustamante, B.; Canton, E.; Fothergill, A.; Fuller, J.; Gonzalez, G.M.; Lass-Flörl, C.; Lockhart, S.R.; Martin-Mazuelos, E.; et al. Multilaboratory study of epidemiological cutoff values for detection of resistance in eight Candida species to fluconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 2014, 58, 2006–2012. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A.; Group, G.A.S. Results from the artemis disk global antifungal surveillance study, 1997 to 2007: A 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, R.; Badiee, P.; Badali, H.; Abastabar, M.; Safa, A.H.; Hadipour, M.; Yazdani, H.; Heshmat, F. Use of restriction fragment length polymorphism to identify Candida species, related to onychomycosis. Adv. Biomed. Res. 2015, 4, 95. [Google Scholar] [PubMed]
- Ghosh, A.K.; Paul, S.; Sood, P.; Rudramurthy, S.M.; Rajbanshi, A.; Jillwin, T.J.; Chakrabarti, A. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the rapid identification of yeasts causing bloodstream infections. Clin. Microbiol. Infect. 2015, 21, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, W.R.; Lopez-Ribot, J.L.; McAtee, R.K.; Patterson, T.F. Growth competition between Candida dubliniensis and Candida albicans under broth and biofilm growing conditions. J. Clin. Microbiol. 2000, 38, 902–904. [Google Scholar] [PubMed]
- Zheng, H.; Kim, J.; Liew, M.; Yan, J.K.; Herrera, O.; Bok, J.W.; Kelleher, N.L.; Keller, N.P.; Wang, Y. Redox metabolites signal polymicrobial biofilm development via the napa oxidative stress cascade in aspergillus. Curr. Biol. 2015, 25, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Manavathu, E.K.; Vager, D.L.; Vazquez, J.A. Development and antimicrobial susceptibility studies of in vitro monomicrobial and polymicrobial biofilm models with Aspergillus fumigatus and Pseudomonas aeruginosa. BMC Microbiol. 2014, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Tampakakis, E.; Peleg, A.Y.; Mylonakis, E. Interaction of Candida albicans with an intestinal pathogen, Salmonella enterica serovar typhimurium. Eukaryot. Cell 2009, 8, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. In vitro growth and analysis of Candida biofilms. Nat. Protoc. 2008, 3, 1909–1924. [Google Scholar] [CrossRef] [PubMed]
- Doll, K.; Jongsthaphongpun, K.L.; Stumpp, N.S.; Winkel, A.; Stiesch, M. Quantifying implant-associated biofilms: Comparison of microscopic, microbiologic and biochemical methods. J. Microbiol. Methods 2016, 130, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Krom, B.P.; Cohen, J.B.; McElhaney Feser, G.E.; Cihlar, R.L. Optimized Candidal biofilm microtiter assay. J. Microbiol. Methods 2007, 68, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Krom, B.P.; Willems, H.M. In vitro models for Candida biofilm development. Methods Mol. Biol. 2016, 1356, 95–105. [Google Scholar] [PubMed]
- Trafny, E.A.; Lewandowski, R.; Zawistowska-Marciniak, I.; Stepinska, M. Use of MTT assay for determination of the biofilm formation capacity of microorganisms in metalworking fluids. World J. Microbiol. Biotechnol. 2013, 29, 1635–1643. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; Balkis, M.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J. Clin. Microbiol. 2003, 41, 506–508. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Casadevall, A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infect. Immun. 2006, 74, 6118–6123. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.M.; Hoogenkamp, M.A.; van der Sluis, L.W.; Wesselink, P.R.; Crielaard, W.; Deng, D.M. Resazurin metabolism assay for root canal disinfectant evaluation on dual-species biofilms. J. Endod. 2011, 37, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Shopova, I.; Bruns, S.; Thywissen, A.; Kniemeyer, O.; Brakhage, A.A.; Hillmann, F. Extrinsic extracellular dna leads to biofilm formation and colocalizes with matrix polysaccharides in the human pathogenic fungus Aspergillus fumigatus. Front. Microbiol. 2013, 4, 141. [Google Scholar] [CrossRef] [PubMed]
- Fessia, S.L.; Griffin, M.J. A method for assaying biofilm capacity on polyurethane-coated slides. Perit. Dial. Int. 1991, 11, 144–146. [Google Scholar] [PubMed]
- Di Bonaventura, G.; Pompilio, A.; Picciani, C.; Iezzi, M.; D’Antonio, D.; Piccolomini, R. Biofilm formation by the emerging fungal pathogen Trichosporon asahii: Development, architecture, and antifungal resistance. Antimicrob. Agents Chemother. 2006, 50, 3269–3276. [Google Scholar] [CrossRef] [PubMed]
- Sohm, J.A.; Edwards, B.R.; Wilson, B.G.; Webb, E.A. Constitutive extracellular polysaccharide (EPS) production by specific isolates of Crocosphaera watsonii. Front. Microbiol. 2011, 2, 229. [Google Scholar] [CrossRef] [PubMed]
- Tote, K.; Vanden Berghe, D.; Maes, L.; Cos, P. A new colorimetric microtitre model for the detection of Staphylococcus aureus biofilms. Lett. Appl. Microbiol. 2008, 46, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Hawser, S.P.; Baillie, G.S.; Douglas, L.J. Production of extracellular matrix by Candida albicans biofilms. J. Med. Microbiol. 1998, 47, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, J.R.; Swerhone, G.D.; Leppard, G.G.; Araki, T.; Zhang, X.; West, M.M.; Hitchcock, A.P. Scanning transmission X-ray, laser scanning, and transmission electron microscopy mapping of the exopolymeric matrix of microbial biofilms. Appl. Environ. Microbiol. 2003, 69, 5543–5554. [Google Scholar] [CrossRef] [PubMed]
- Priester, J.H.; Horst, A.M.; Van de Werfhorst, L.C.; Saleta, J.L.; Mertes, L.A.; Holden, P.A. Enhanced visualization of microbial biofilms by staining and environmental scanning electron microscopy. J. Microbiol. Methods 2007, 68, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Bridier, A.; Meylheuc, T.; Briandet, R. Realistic representation of Bacillus subtilis biofilms architecture using combined microscopy (CLSM, ESEM and FESEM). Micron 2013, 48, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Dige, I.; Nilsson, H.; Kilian, M.; Nyvad, B. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur. J. Oral Sci. 2007, 115, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Ivleva, N.P.; Haisch, C.; Niessner, R.; Horn, H. Combined use of confocal laser scanning microscopy (CLSM) and raman microscopy (RM): Investigations on EPS-Matrix. Water Res. 2009, 43, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Surman, S.B.; Walker, J.T.; Goddard, D.T.; Morton, L.H.G.; Keevil, C.W.; Weaver, W.; Skinner, A.; Hanson, K.; Caldwell, D.; Kurtz, J. Comparison of microscope techniques for the examination of biofilms. J. Microbiol. Methods 1996, 25, 57–70. [Google Scholar] [CrossRef]
- Repp, K.K.; Menor, S.A.; Pettit, R.K. Microplate alamar blue assay for susceptibility testing of Candida albicans biofilms. Med. Mycol. 2007, 45, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K.; Weber, C.A.; Kean, M.J.; Hoffmann, H.; Pettit, G.R.; Tan, R.; Franks, K.S.; Horton, M.L. Microplate alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob. Agents Chemother. 2005, 49, 2612–2617. [Google Scholar] [CrossRef] [PubMed]
- Muszkieta, L.; Beauvais, A.; Pahtz, V.; Gibbons, J.G.; Anton Leberre, V.; Beau, R.; Shibuya, K.; Rokas, A.; Francois, J.M.; Kniemeyer, O.; et al. Investigation of Aspergillus fumigatus biofilm formation by various “Omics” approaches. Front. Microbiol. 2013, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, C.L.; Mann, M. Proteomics. Annu. Rev. Genom. Hum. Genet. 2004, 5, 267–293. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, N.F.; Lopes, S.P.; Keevil, C.W.; Pereira, M.O.; Vieira, M.J. Time to “Go large” On biofilm research: Advantages of an omics approach. Biotechnol. Lett. 2009, 31, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; May, A.; Sherry, L.; Kean, R.; Williams, C.; Jones, B.L.; Burgess, K.V.; Heringa, J.; Abeln, S.; Brandt, B.W.; et al. Integrating Candida albicans metabolism with biofilm heterogeneity by transcriptome mapping. Sci. Rep. 2016, 6, 35436. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Bernhardt, J.; Hecker, M.; Becher, D. Global relative and absolute quantitation in microbial proteomics. Curr. Opin. Microbiol. 2012, 15, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.P.; Bachmann, S.P.; Lopez-Ribot, J.L. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics 2006, 6, 5795–5804. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, C.J.; Wang, Y.; Jin, L.; Abiko, Y.; Samaranayake, L.P. Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 2008, 8, 2936–2947. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gomariz, M.; Perumal, P.; Mekala, S.; Nombela, C.; Chaffin, W.L.; Gil, C. Proteomic analysis of cytoplasmic and surface proteins from yeast cells, hyphae, and biofilms of Candida albicans. Proteomics 2009, 9, 2230–2252. [Google Scholar] [CrossRef] [PubMed]
- Santi, L.; Beys-da-Silva, W.O.; Berger, M.; Calzolari, D.; Guimaraes, J.A.; Moresco, J.J.; Yates, J.R., 3rd. Proteomic profile of Cryptococcus neoformans biofilm reveals changes in metabolic processes. J. Proteome Res. 2014, 13, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Buescher, J.M.; Driggers, E.M. Integration of omics: More than the sum of its parts. Cancer Metab. 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Chaturvedi, A.K.; Lopez-Ribot, J.L. Design of a simple model of Candida albicans biofilms formed under conditions of flow: Development, architecture, and drug resistance. Mycopathologia 2009, 168, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Andes, D.R. Fungal biofilms: In vivo models for discovery of anti-biofilm drugs. Microbiol. Spectr. 2015, 3, E30. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, N.; Li, D.Q.; Ljungh, A. Protein adsorption on ex vivo catheters and polymers exposed to peritoneal dialysis effluent. Perit. Dial. Int. 2004, 24, 264–273. [Google Scholar] [PubMed]
- Dongari-Bagtzoglou, A. Mucosal biofilms: Challenges and future directions. Expert Rev. Anti Infect. Ther. 2008, 6, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Casadevall, A. Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Appl. Environ. Microbiol. 2007, 73, 4592–4601. [Google Scholar] [CrossRef] [PubMed]
- Ronsani, M.M.; Mores Rymovicz, A.U.; Meira, T.M.; Trindade Grégio, A.M.; Guariza Filho, O.; Tanaka, O.M.; Ribeiro Rosa, E.A. Virulence modulation of Candida albicans biofilms by metal ions commonly released from orthodontic devices. Microb. Pathog. 2011, 51, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; McCormick, T.S.; Imamura, Y.; Mukherjee, P.K.; Ghannoum, M.A. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. Albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect. Immun. 2007, 75, 2612–2620. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Casadevall, A. Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect. Immun. 2005, 73, 6350–6362. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms and device-associated infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Lilly, E.A.; Rodriguez, T.E.; Fidel, P.L.; Noverr, M.C. Candida albicans forms biofilms on the vaginal mucosa. Microbiology 2010, 156, 3635–3644. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.R. In Vivo Candida Device Biofilm Models. In Candida albicans: Cellular and Molecular Biology; Prasad, R., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 93–113. [Google Scholar]
- Ghannoum, M.; Roilides, E.; Katragkou, A.; Petraitis, V.; Walsh, T.J. The role of echinocandins in Candida biofilm-related vascular catheter infections: In vitro and in vivo model systems. Clin. Infect. Dis. 2015, 61, S618–S621. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.; Nett, J.; Oschel, P.; Albrecht, R.; Marchillo, K.; Pitula, A. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect. Immun. 2004, 72, 6023–6031. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.R.; Mihu, M.R.; Tar, M.; Cordero, R.J.; Han, G.; Friedman, A.J.; Friedman, J.M.; Nosanchuk, J.D. Demonstration of antibiofilm and antifungal efficacy of chitosan against Candidal biofilms, using an in vivo central venous catheter model. J. Infect. Dis. 2010, 201, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Schinabeck, M.K.; Long, L.A.; Hossain, M.A.; Chandra, J.; Mukherjee, P.K.; Mohamed, S.; Ghannoum, M.A. Rabbit model of Candida albicans biofilm infection: Liposomal amphotericin B antifungal lock therapy. Antimicrob. Agents Chemother. 2004, 48, 1727–1732. [Google Scholar] [CrossRef] [PubMed]
- Kucharikova, S.; Neirinck, B.; Sharma, N.; Vleugels, J.; Lagrou, K.; van Dijck, P. In vivo Candida glabrata biofilm development on foreign bodies in a rat subcutaneous model. J. Antimicrob. Chemother. 2015, 70, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Sangeorzan, J.A.; Bradley, S.F.; He, X.; Zarins, L.T.; Ridenour, G.L.; Tiballi, R.N.; Kauffman, C.A. Epidemiology of oral candidiasis in HIV-infected patients: Colonization, infection, treatment, and emergence of fluconazole resistance. Am. J. Med. 1994, 97, 339–346. [Google Scholar] [CrossRef]
- Solis, N.V.; Filler, S.G. Mouse model of oropharyngeal candidiasis. Nat. Protoc. 2012, 7, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Marchillo, K.; Spiegel, C.A.; Andes, D.R. Development and validation of an in vivo Candida albicans biofilm denture model. Infect. Immun. 2010, 78, 3650–3659. [Google Scholar] [CrossRef] [PubMed]
- Freire, F.; Ferraresi, C.; Jorge, A.O.; Hamblin, M.R. Photodynamic therapy of oral Candida infection in a mouse model. J. Photochem. Photobiol. B 2016, 159, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Seleem, D.; Benso, B.; Noguti, J.; Pardi, V.; Murata, R.M. In vitro and in vivo antifungal activity of lichochalcone-a against Candida albicans biofilms. PLoS ONE 2016, 11, e0157188. [Google Scholar] [CrossRef] [PubMed]
- Vila, T.V.; Chaturvedi, A.K.; Rozental, S.; Lopez-Ribot, J.L. In vitro activity of miltefosine against Candida albicans under planktonic and biofilm growth conditions and in vivo efficacy in a murine model of oral candidiasis. Antimicrob. Agents Chemother. 2015, 59, 7611–7620. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fries, B.C. A murine model for catheter-associated candiduria. J. Med. Microbiol. 2011, 60, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Brooks, E.G.; Cabezas-Olcoz, J.; Sanchez, H.; Zarnowski, R.; Marchillo, K.; Andes, D.R. Rat indwelling urinary catheter model of Candida albicans biofilm infection. Infect. Immun. 2014, 82, 4931–4940. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chandra, J.; Mukherjee, P.; Szczotka-Flynn, L.; Ghannoum, M.A.; Pearlman, E. A murine model of contact lens-associated Fusarium keratitis. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Pinnock, A.; Shivshetty, N.; Roy, S.; Rimmer, S.; Douglas, I.; MacNeil, S.; Garg, P. Ex vivo rabbit and human corneas as models for bacterial and fungal keratitis. Graefes Arch. Clin. Exp. Ophthalmol. 2017, 255, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, L.; Li, Z.; Liu, S.; Xie, Y.; He, S.; Deng, X.; Yang, B.; Liu, H.; Chen, G.; et al. A novel murine model of Fusarium solani keratitis utilizing fluorescent labeled fungi. Exp. Eye Res. 2013, 110, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Benaducci, T.; Sardi Jde, C.; Lourencetti, N.M.; Scorzoni, L.; Gullo, F.P.; Rossi, S.A.; Derissi, J.B.; de Azevedo Prata, M.C.; Fusco-Almeida, A.M.; Mendes-Giannini, M.J. Virulence of Cryptococcus sp. Biofilms in vitro and in vivo using Galleria mellonella as an alternative model. Front. Microbiol. 2016, 7, 290. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R.; Borghi, E.; Falleni, M.; Perdoni, F.; Tosi, D.; Lappin, D.F.; O'Donnell, L.; Greetham, D.; Ramage, G.; Nile, C. Acetylcholine protects against Candida albicans infection by inhibiting biofilm formation and promoting hemocyte function in a Galleria mellonella infection model. Eukaryot. Cell 2015, 14, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Cirasola, D.; Sciota, R.; Vizzini, L.; Ricucci, V.; Morace, G.; Borghi, E. Experimental biofilm-related Candida infections. Future Microbiol. 2013, 8, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. The extracellular matrix of fungal biofilms. Adv. Exp. Med. Biol. 2016, 931, 21–35. [Google Scholar] [PubMed]
- Mathe, L.; van Dijck, P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr. Genet. 2013, 59, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Sardi, J.; Pitangui, N.; Gullo, F.; Fusco-Almeida, A.; Mendes-Giannini, A. Fungal biofilms: Formation, resistance and pathogenicity. In Medical Mycology: Current Trends and Future Prospects; Razzaghi-Abyaneh, M., Shams-Ghahfarokhi, M., Rai, M., Eds.; Taylor & Francis Group: Oxford, UK, 2015; Volume 1, pp. 291–314. [Google Scholar]
- Perumal, P.; Mekala, S.; Chaffin, W.L. Role for cell density in antifungal drug resistance in Candida albicans biofilms. Antimicrob. Agents Chemother. 2007, 51, 2454–2463. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, C.J.; Jin, L.; Samaranayake, L.P. Biofilm lifestyle of candida: A mini review. Oral Dis. 2008, 14, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Nailis, H.; Vandenbosch, D.; Deforce, D.; Nelis, H.J.; Coenye, T. Transcriptional response to fluconazole and amphotericin b in Candida albicans biofilms. Res. Microbiol. 2010, 161, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E. Future directions for anti-biofilm therapeutics targeting candida. Expert Rev. Anti Infect. Ther. 2014, 12, 375–382. [Google Scholar] [CrossRef] [PubMed]
- LaFleur, M.D.; Kumamoto, C.A.; Lewis, K. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob. Agents Chemother. 2006, 50, 3839–3846. [Google Scholar] [CrossRef] [PubMed]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Diez-Orejas, R.; Molero, G.; Navarro-Garcia, F.; Pla, J.; Nombela, C.; Sanchez-Perez, M. Reduced virulence of Candida albicans mkc1 mutants: A role for mitogen-activated protein kinase in pathogenesis. Infect. Immun. 1997, 65, 833–837. [Google Scholar] [PubMed]
- Mowat, E.; Lang, S.; Williams, C.; McCulloch, E.; Jones, B.; Ramage, G. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J. Antimicrob. Chemother. 2008, 62, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Niimi, M.; Firth, N.A.; Cannon, R.D. Antifungal drug resistance of oral fungi. Odontology 2010, 98, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Rajendran, R.; Sherry, L.; Williams, C. Fungal biofilm resistance. Int. J. Microbiol. 2012, 2012, 528521. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Stevens, D.A.; Cegelski, L. Fungal biofilm composition and opportunities in drug discovery. Future Med. Chem. 2016, 8, 1455–1468. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Mitchell, K.F.; Edward, J.A.; Andes, D.R. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013, 8, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Mowat, E.; Jones, B.; Williams, C.; Lopez-Ribot, J. Our current understanding of fungal biofilms. Crit. Rev. Microbiol. 2009, 35, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Wongsuk, T.; Pumeesat, P.; Luplertlop, N. Fungal quorum sensing molecules: Role in fungal morphogenesis and pathogenicity. J. Basic Microbiol. 2016, 56, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi—A review. Med. Mycol. 2012, 50, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; Lopez-Ribot, J.L. Candida biofilms: An update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Whiteway, M. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell 2005, 4, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Prasad, R. The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans. Antimicrob. Agents Chemother. 2011, 55, 4834–4843. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Saville, S.P.; Wickes, B.L.; Lopez-Ribot, J.L. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 2002, 68, 5459–5463. [Google Scholar] [CrossRef] [PubMed]
- Semighini, C.P.; Hornby, J.M.; Dumitru, R.; Nickerson, K.W.; Harris, S.D. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 2006, 59, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Jabra-Rizk, M.A.; Shirtliff, M.; James, C.; Meiller, T. Effect of farnesol on Candida dubliniensis biofilm formation and fluconazole resistance. FEMS Yeast Res. 2006, 6, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Persister cells: Molecular mechanisms related to antibiotic tolerance. Handb. Exp. Pharmacol. 2012, 121–133. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J.; Kuhn, D.M.; Ghannoum, M.A. Mechanism of fluconazole resistance in Candida albicans biofilms: Phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 2003, 71, 4333–4340. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, K.W.; Atkin, A.L.; Hornby, J.M. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl. Environ. Microbiol. 2006, 72, 3805–3813. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Henriques, M.; Azeredo, J.; Rocha, S.M.; Coimbra, M.A.; Oliveira, R. Morphogenesis control in Candida albicans and Candida dubliniensis through signaling molecules produced by planktonic and biofilm cells. Eukaryot. Cell 2007, 6, 2429–2436. [Google Scholar] [CrossRef] [PubMed]
- Shirtliff, M.E.; Krom, B.P.; Meijering, R.A.; Peters, B.M.; Zhu, J.; Scheper, M.A.; Harris, M.L.; Jabra-Rizk, M.A. Farnesol-induced apoptosis in Candida albicans. Antimicrob. Agents Chemother. 2009, 53, 2392–2401. [Google Scholar] [CrossRef] [PubMed]
- Alem, M.A.; Oteef, M.D.; Flowers, T.H.; Douglas, L.J. Production of tyrosol by Candida albicans biofilms and its role in quorum sensing and biofilm development. Eukaryot. Cell 2006, 5, 1770–1779. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fink, G.R. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006, 20, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro Rde, A.; Teixeira, C.E.; Brilhante, R.S.; Castelo-Branco, D.S.; Alencar, L.P.; de Oliveira, J.S.; Monteiro, A.J.; Bandeira, T.J.; Sidrim, J.J.; Moreira, J.L.; et al. Exogenous tyrosol inhibits planktonic cells and biofilms of Candida species and enhances their susceptibility to antifungals. FEMS Yeast Res. 2015, 15, fov012. [Google Scholar] [CrossRef] [PubMed]
- Lorek, J.; Poggeler, S.; Weide, M.R.; Breves, R.; Bockmuhl, D.P. Influence of farnesol on the morphogenesis of Aspergillus niger. J. Basic Microbiol. 2008, 48, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Dichtl, K.; Ebel, F.; Dirr, F.; Routier, F.H.; Heesemann, J.; Wagener, J. Farnesol misplaces tip-localized ρ proteins and inhibits cell wall integrity signalling in Aspergillus fumigatus. Mol. Microbiol. 2010, 76, 1191–1204. [Google Scholar] [CrossRef] [PubMed]
- Brilhante, R.S.; de Lima, R.A.; Marques, F.J.; Silva, N.F.; Caetano, E.P.; Castelo-Branco Dde, S.; Bandeira Tde, J.; Moreira, J.L.; Cordeiro Rde, A.; Monteiro, A.J.; et al. Histoplasma capsulatum in planktonic and biofilm forms: In vitro susceptibility to amphotericin B, itraconazole and farnesol. J. Med. Microbiol. 2015, 64, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Derengowski, L.S.; De-Souza-Silva, C.; Braz, S.V.; Mello-De-Sousa, T.M.; Bao, S.N.; Kyaw, C.M.; Silva-Pereira, I. Antimicrobial effect of farnesol, a Candida albicans quorum sensing molecule, on Paracoccidioides brasiliensis growth and morphogenesis. Ann. Clin. Microbiol. Antimicrob. 2009, 8, 13. [Google Scholar] [CrossRef] [PubMed]
| Microtiter Plates Assays | Characteristics |
|---|---|
| MTT | MTT is a yellow soluble salt, which in the presence of metabolic activity, is reduced to an insoluble purple formazan crystal. This method is used to determine the metabolic activity of some microorganisms in planktonic and biofilm forms. Moreover, this method shows excellent correlation with biomass determination by dry weight. Fast and convenient [24]. |
| XTT | Tetrazolium salt (yellow) is reduced by the activity of fungal mitochondrial dehydrogenase to formazan salt (orange), which is correlated with cell viability. It is also used to determine metabolic activity in the developmental stages of biofilms and in antifungal susceptibility tests [46,116,118]. The method is simple and, reproducible, but some disadvantages were reported by Khun et al. [116]. |
| Alamar Blue and Resazurin | Reduction is dependent on metabolic activity. The methods are fast and simple and measurement can be conducted spectrofluorometrically or spectrophotometrically. Resazurin is the active principle of Alamar Blue. The reagents are nontoxic to humans and fungi and the method is reproducible. Good correlation with XTT assay and CFU/mL [133,134]. Used for biofilm quantification. Blue dye resazurin is converted to pink resorufin in the presence of metabolic activity. Nontoxic and soluble in water [119,120,121]. |
| Safranin | Dye easy to use for ECM quantification Difficult interpretation; low-cost [7,42,122]. |
| Crystal Violet (CV) | Used for biomass quantification. CV stains living and dead cells, and thus it is not indicated to verify antifungal activity in biofilms [119]. Low cost and easy [31]. |
| Alcian Blue | Measures mass quantity of biofilm ECM [123,124]. |
| 1,9-Dimethyl Methylene Blue (DMMB) | Quantification of biofilm matrix [119,125]. |
| Resistance Mechanisms | Effect | References |
|---|---|---|
| Cellular density | Quorum sensing | Perumal et al. [180]; Seneviratne et al. [181]. |
| Differential regulation drug target | Alteration in target levels; Associated with changes in target structure that make the drug unable to bind to the target. | Nailis et al. [182]. |
| Upregulation drug efflux pumps | Antifungal is pumped out of cells and thus cannot perform its intracellular function. | Nett et al. [183] |
| Persister cells | Because of the dormant state of the persisters, antifungal targets are inactive. | LaFleur et al. [184] |
| Presence of a matrix | Specific binding of antifungals to β-1,3-glucans, a major component of the matrix, prevents antifungal agents from reaching their targets. | Al-Fattani and Douglas [185]; Mitchell et al. [177]. |
| Diverse stress responses | Possible indirect effects through the regulation of other resistance mechanisms. | Diez-Orejas et al. [186] |
| Organism | QSMs | Role of QSMs in Molds and Dimorphic Fungi | References |
|---|---|---|---|
| C. albicans | Farnesol | Inhibited hyphal development Involved in morphogenesis Inhibited biofilm formation Induced apoptosis Antifungal activity Modulated drug extrusion | Nickerson et al. [207] |
| Martins et al. [208] | |||
| Ramage et al. [200] | |||
| Shirtliff et al. [209] | |||
| Sardi et al. [10] | |||
| Sharma et al. [199] | |||
| Tyrosol | Promoted germ tube formation Stimulated hypha production during the early stages of biofilm development Antifungal activity | Alem et al. [210] | |
| Chen et al. [211] | |||
| Cordeiro et al. [212] | |||
| A. niger | Farnesol | Inhibited conidiation Reduced intracellular cAMP levels | Lorek et al. [213] |
| A. fumigatus | Farnesol | Altered growth phenotype Perturbed cell wall | Dichtl et al. [214] |
| H. capsulatum | Farnesol | Inhibited biofilm formation Antifungal activity | Brilhante et al. [215] |
| P. brasiliensis | Farnesol | Inhibited growth Delayed the dimorphic transition Antifungal activity | Derengowski et al. [216] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; De Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcón, K.P.; Melo, W.C.M.A.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal Biofilms and Polymicrobial Diseases. J. Fungi 2017, 3, 22. https://doi.org/10.3390/jof3020022
Costa-Orlandi CB, Sardi JCO, Pitangui NS, De Oliveira HC, Scorzoni L, Galeane MC, Medina-Alarcón KP, Melo WCMA, Marcelino MY, Braz JD, et al. Fungal Biofilms and Polymicrobial Diseases. Journal of Fungi. 2017; 3(2):22. https://doi.org/10.3390/jof3020022
Chicago/Turabian StyleCosta-Orlandi, Caroline B., Janaina C. O. Sardi, Nayla S. Pitangui, Haroldo C. De Oliveira, Liliana Scorzoni, Mariana C. Galeane, Kaila P. Medina-Alarcón, Wanessa C. M. A. Melo, Mônica Y. Marcelino, Jaqueline D. Braz, and et al. 2017. "Fungal Biofilms and Polymicrobial Diseases" Journal of Fungi 3, no. 2: 22. https://doi.org/10.3390/jof3020022