Metabolic Interactions between Bacteria and Fungi in Commensal Oral Biofilms
Abstract
:1. Introduction
1.1. Importance of Oral Health
1.2. Oral Health Is Much More than Healthy Teeth
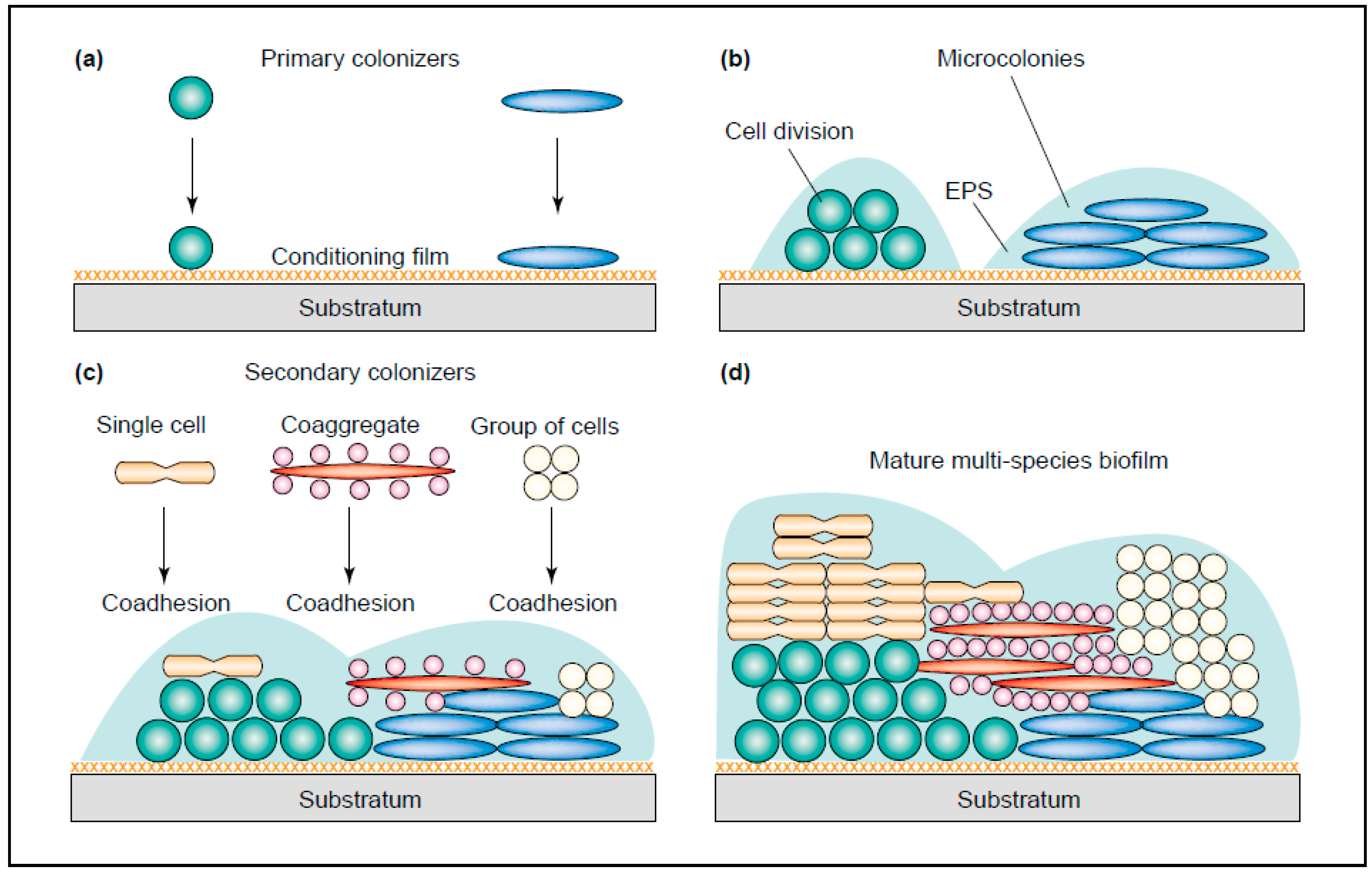
1.3. Oral Microbiota Is Complex
1.4. Bacteria
1.5. Diseases of the Mouth
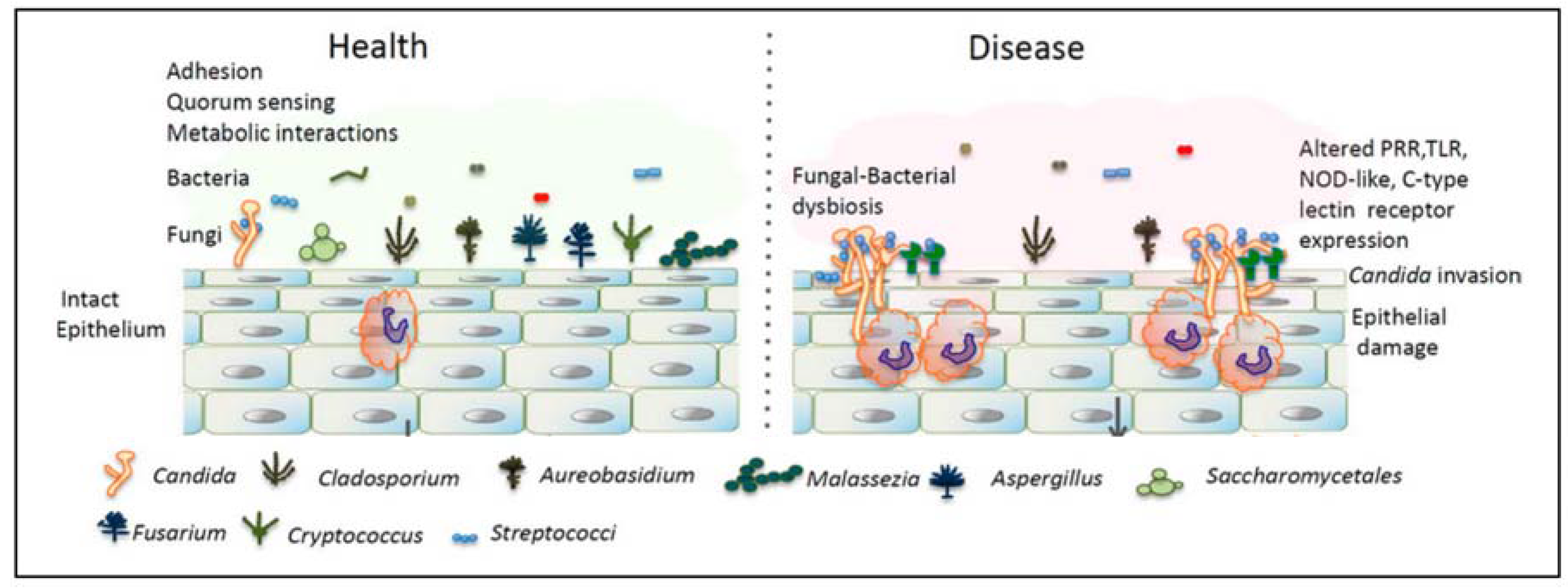
1.6. Fungi
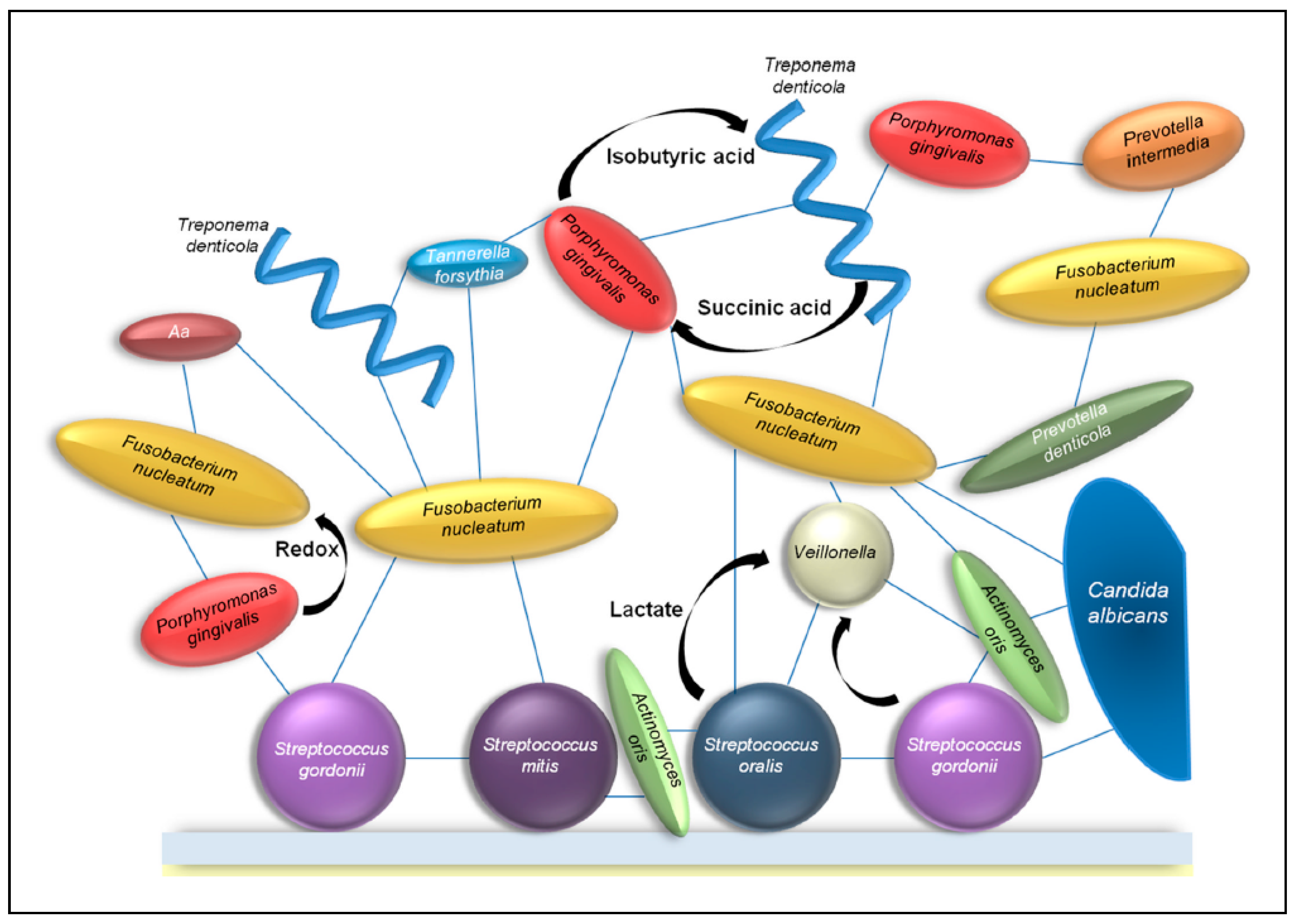
2. Interactions between Fungi and Bacteria Might Be Important for Maintenance of the Healthy Oral Ecology
2.1. Volatile Sulfur Compounds
3. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kleinman, D.V. The future of the dental profession: Perspectives from oral health in America: A report of the surgeon general. J. Am. Coll. Dent. 2002, 69, 6–10. [Google Scholar] [PubMed]
- Larsen, F.J.; Ekblom, B.; Sahlin, K.; Lundberg, J.O.; Weitzberg, E. Effects of dietary nitrate on blood pressure in healthy volunteers. N. Engl. J. Med. 2006, 355, 2792–2793. [Google Scholar] [CrossRef] [PubMed]
- Govoni, M.; Jansson, E.A.; Weitzberg, E.; Lundberg, J.O. The increase in plasma nitrite after a dietary nitrate load is markedly attenuated by an antibacterial mouthwash. Nitric Oxide 2008, 19, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Filoche, S.; Wong, L.; Sissons, C.H. Oral biofilms: Emerging concepts in microbial ecology. J. Dent. Res. 2010, 89, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Kanasi, E.; Dewhirst, F.E.; Chalmers, N.I.; Kent, R., Jr.; Moore, A.; Hughes, C.V.; Pradhan, N.; Loo, C.Y.; Tanner, A.C. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010, 44, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Rosier, B.T.; De Jager, M.; Zaura, E.; Krom, B.P. Historical and contemporary hypotheses on the development of oral diseases: Are we there yet? Front. Cell. Infect. Microbiol. 2014, 4, 92. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, H.F.; Lappin-Scott, H.M. Biofilms adhere to stay. Trends Microbiol. 2001, 9, 9–10. [Google Scholar] [CrossRef]
- Van Leeuwenhoek, A. About animals in the scrurf of the teeth. Philos. Trans. R. Soc. Lond. 1683, 14, 568–574. [Google Scholar]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Gomez, A.; Nelson, K.E. The oral microbiome of children: Development, disease, and implications beyond oral health. Microb. Ecol. 2017, 73, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Keijser, B.J.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Zarco, M.F.; Vess, T.J.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Simon-Soro, A.; Tomas, I.; Cabrera-Rubio, R.; Catalan, M.D.; Nyvad, B.; Mira, A. Microbial geography of the oral cavity. J. Dent. Res. 2013, 92, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Hannig, M. Ultrastructural investigation of pellicle morphogenesis at two different intraoral sites during a 24 h period. Clin. Oral Investig. 1999, 3, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E.; Andersen, R.N.; Blehert, D.S.; Egland, P.G.; Foster, J.S.; Palmer, R.J., Jr. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 2002, 66, 486–505. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C. Microbiology: Breaking down biofilms. Curr. Biol. 2002, 12, R132–R134. [Google Scholar] [CrossRef]
- Rickard, A.H.; Gilbert, P.; High, N.J.; Kolenbrander, P.E.; Handley, P.S. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003, 11, 94–100. [Google Scholar] [CrossRef]
- Kleinberg, I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: An alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit. Rev. Oral Biol. Med. 2002, 13, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Beighton, D. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent. Oral Epidemiol. 2005, 33, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W. Dental caries and periodontitis: Contrasting two infections that have medical implications. Infect. Dis. Clin. N. Am. 2007, 21, 471–502. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.B.; Karpinia, K.; Baehni, P. Chemotherapeutics: Antibiotics and other antimicrobials. Periodontology 2000 2004, 36, 146–165. [Google Scholar] [CrossRef] [PubMed]
- Aylikci, B.U.; Colak, H. Halitosis: From diagnosis to management. J. Nat. Sci. Biol. Med. 2013, 4, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. Microbiology and treatment of halitosis. Curr. Infect. Dis. Rep. 2003, 5, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Krespi, Y.P.; Shrime, M.G.; Kacker, A. The relationship between oral malodor and volatile sulfur compound-producing bacteria. Otolaryngol. Head Neck Surg. 2006, 135, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Lucas, V.S.; Gafan, G.; Dewhurst, S.; Roberts, G.J. Prevalence, intensity and nature of bacteraemia after toothbrushing. J. Dent. 2008, 36, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, P.B.; Brennan, M.T.; Thornhill, M.; Michalowicz, B.S.; Noll, J.; Bahrani-Mougeot, F.K.; Sasser, H.C. Poor oral hygiene as a risk factor for infective endocarditis-related bacteremia. J. Am. Dent. Assoc. 2009, 140, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.J.; Ford, P.J.; Cullinan, M.P.; Leishman, S.; Yamazaki, K. Relationship between periodontal infections and systemic disease. Clin. Microbiol. Infect. 2007, 13, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Kinane, D.; Bouchard, P. Periodontal diseases and health: Consensus report of the sixth European workshop on periodontology. J. Clin. Periodontol. 2008, 35, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. Plaque as a biofilm: Pharmacological principles of drug delivery and action in the sub- and supragingival environment. Oral Dis. 2003, 9, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.D.; Underhill, D.M. Striking a balance: Fungal commensalism versus pathogenesis. Curr. Opin. Microbiol. 2013, 16, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Bowman, B.H.; Taylor, J.W.; White, T.J. Molecular evolution of the fungi: Human pathogens. Mol. Biol. Evol. 1992, 9, 893–904. [Google Scholar] [PubMed]
- Cooney, N.M.; Klein, B.S. Fungal adaptation to the mammalian host: It is a new world, after all. Curr. Opin. Microbiol. 2008, 11, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-da-Silva, F.; Araujo, R.; Sampaio-Maia, B. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med. Mycol. 2014, 52, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.S.; Lynch, N.; McCullough, M.J. Oral fungal infections: An update for the general practitioner. Aust. Dent. J. 2010, 55 (Suppl. 1), 48–54. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Dongari-Bagtzoglou, A. Shaping the oral mycobiota: Interactions of opportunistic fungi with oral bacteria and the host. Curr. Opin. Microbiol. 2015, 26, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Kolenbrander, P.E. Oral microbial communities: Biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 2000, 54, 413–437. [Google Scholar] [CrossRef] [PubMed]
- Zijnge, V.; van Leeuwen, M.B.; Degener, J.E.; Abbas, F.; Thurnheer, T.; Gmur, R.; Harmsen, H.J. Oral biofilm architecture on natural teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef] [PubMed]
- Janus, M.M.; Willems, H.M.; Krom, B.P. Candida albicans in multispecies oral communities; a keystone commensal? Adv. Exp. Med. Biol. 2016, 931, 13–20. [Google Scholar] [PubMed]
- Wright, C.J.; Burns, L.H.; Jack, A.A.; Back, C.R.; Dutton, L.C.; Nobbs, A.H.; Lamont, R.J.; Jenkinson, H.F. Microbial interactions in building of communities. Mol. Oral Microbiol. 2013, 28, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, L.M.; Deng, D.M.; van der Mei, H.C.; Crielaard, W.; Krom, B.P. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot. Cell 2009, 8, 1658–1664. [Google Scholar] [CrossRef] [PubMed]
- Bachtiar, E.W.; Bachtiar, B.M.; Jarosz, L.M.; Amir, L.R.; Sunarto, H.; Ganin, H.; Meijler, M.M.; Krom, B.P. Ai-2 of aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front. Cell. Infect. Microbiol. 2014, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Hogan, D.A.; Vik, A.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef] [PubMed]
- Bamford, C.V.; d’Mello, A.; Nobbs, A.H.; Dutton, L.C.; Vickerman, M.M.; Jenkinson, H.F. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 2009, 77, 3696–3704. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Long, C.D.; Armitage, G.C.; Loomer, P.; Emerson, J.; Mongodin, E.F.; Nelson, K.E.; Gill, S.R.; Fraser-Liggett, C.M.; Relman, D.A. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010, 4, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Egland, P.G.; Robert, J.; Palmer, J.; Kolenbrander, P.E. Interspecies communication in Streptococcus gordonii–Veillonella atypica biofilms: Signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 2004, 101, 16917–16922. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D.; Martin, M.V. Oral Microbiology, 5th ed.; Elsevier Health Sciences: London, UK, 2009; p. 232. [Google Scholar]
- Han, T.L.; Cannon, R.D.; Villas-Boas, S.G. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet. Biol. 2011, 48, 747–763. [Google Scholar] [CrossRef] [PubMed]
- Metwalli, K.H.; Khan, S.A.; Krom, B.P.; Jabra-Rizk, M.A. Streptococcus mutans, Candida albicans, and the human mouth: A sticky situation. PLoS Pathog. 2013, 9, e1003616. [Google Scholar] [CrossRef] [PubMed]
- Krom, B.P.; Kidwai, S.; Ten Cate, J.M. Candida and other fungal species: Forgotten players of healthy oral microbiota. J. Dent. Res. 2014, 93, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Vylkova, S.; Carman, A.J.; Danhof, H.A.; Collette, J.R.; Zhou, H.; Lorenz, M.C. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. Mbio 2011, 2, e00055-11. [Google Scholar] [CrossRef] [PubMed]
- Danhof, H.A.; Vylkova, S.; Vesely, E.M.; Ford, A.E.; Gonzalez-Garay, M.; Lorenz, M.C. Robust extracellular pH modulation by Candida albicans during growth in carboxylic acids. Mbio 2016, 7, e01646-16. [Google Scholar] [CrossRef] [PubMed]
- Alkan, N.; Fluhr, R.; Sherman, A.; Prusky, D. Role of ammonia secretion and pH modulation on pathogenicity of Colletotrichum coccodes on tomato fruit. Mol. Plant Microbe Interact. 2008, 21, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Morales, D.K.; Hogan, D.A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010, 6, e1000886. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Janus, M.M.; Crielaard, W.; Volgenant, C.M.; van der Veen, M.H.; Brandt, B.W.; Krom, B.P. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 2017, 9, 1270613. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, P.T.; van der Peet, J.M.; Bikker, F.J.; Hoogenkamp, M.A.; Oliveira Paiva, A.M.; Kostidis, S.; Mayboroda, O.A.; Smits, W.K.; Krom, B.P. Interspecies interactions between Clostridium difficile and Candida albicans. mSphere 2016, 1, e00187-16. [Google Scholar] [CrossRef] [PubMed]
- Lachke, S.A.; Lockhart, S.R.; Daniels, K.J.; Soll, D.R. Skin facilitates Candida albicans mating. Infect. Immun. 2003, 71, 4970–4976. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Cao, C.; Liang, W.; Guan, G.; Zhang, Q.; Nobile, C.J.; Huang, G. White cells facilitate opposite- and same-sex mating of opaque cells in Candida albicans. PLoS Genet. 2014, 10, e1004737. [Google Scholar] [CrossRef] [PubMed]
- Amich, J.; Schafferer, L.; Haas, H.; Krappmann, S. Regulation of sulphur assimilation is essential for virulence and affects iron homeostasis of the human-pathogenic mould Aspergillus fumigatus. PLoS Pathog. 2013, 9, e1003573. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.K.; Desai, P.R.; Rai, M.N.; Kaur, R.; Ganesan, K.; Bachhawat, A.K. Glutathione biosynthesis in the yeast pathogens Candida glabrata and Candida albicans: Essential in C. Glabrata., and essential for virulence in C. Albicans. Microbiology 2011, 157, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Marzluf, G.A. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu. Rev. Microbiol. 1997, 51, 73–96. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lof, M.; Janus, M.M.; Krom, B.P. Metabolic Interactions between Bacteria and Fungi in Commensal Oral Biofilms. J. Fungi 2017, 3, 40. https://doi.org/10.3390/jof3030040
Lof M, Janus MM, Krom BP. Metabolic Interactions between Bacteria and Fungi in Commensal Oral Biofilms. Journal of Fungi. 2017; 3(3):40. https://doi.org/10.3390/jof3030040
Chicago/Turabian StyleLof, Marloes, Marleen M. Janus, and Bastiaan P. Krom. 2017. "Metabolic Interactions between Bacteria and Fungi in Commensal Oral Biofilms" Journal of Fungi 3, no. 3: 40. https://doi.org/10.3390/jof3030040



