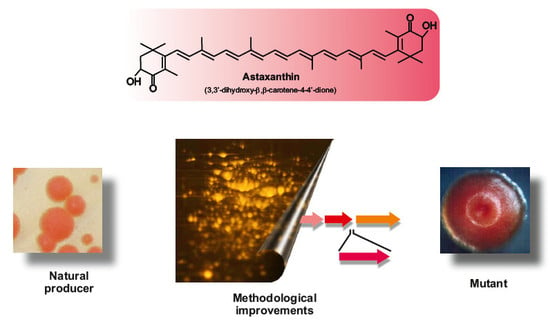
Biosynthesis of Astaxanthin as a Main Carotenoid in the Heterobasidiomycetous Yeast Xanthophyllomyces dendrorhous
Abstract
:1. Introduction: Xanthophyllomyces dendrorhous and Carotenoids
2. The Biosynthetic Pathway of Astaxanthin in X. dendrorhous
3. Biotechnology-Based Improvement of Astaxanthin Production in X. dendrorhous
4. Omics of X. dendrorhous: Genomics, Transcriptomics, Proteomics, and Metabolomics
5. Conclusions and Future Prospects
Acknowledgments
Conflicts of Interest
References
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. In Current Protocols Food Analytical Chemistry; John Wiley & Sons, Inc.: Franklin, NJ, USA, 2001. [Google Scholar] [CrossRef]
- Sandmann, G.; Misawa, N. Fungal Carotenoids. In Industrial Application; Osiewacz, H.D., Ed.; Springer: Berlin/Heidelberg, Germany, 2002; pp. 247–262. [Google Scholar] [CrossRef]
- Sieiro, C.; Poza, M.; de Miguel, T.; Villa, T.G. Genetic basis of microbial carotenogenesis. Int. Microbiol. 2003, 6, 11–16. [Google Scholar] [PubMed]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef] [PubMed]
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. Carotenoids; Birkhäuser Basel: Basel, Switzerland, 2004. [Google Scholar] [CrossRef]
- Bhosale, P.; Bernstein, P.S. Microbial xanthophylls. Appl. Microbiol. Biotechnol. 2005, 68, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Young, A.J. The photoprotective role of carotenoids in higher plants. Physiol. Plant. 1991, 83, 702–708. [Google Scholar] [CrossRef]
- Blomhoff, R.; Blomhoff, H.K. Overview of retinoid metabolism and function. J. Neurobiol. 2006, 66, 606–630. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef] [PubMed]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef] [PubMed]
- Biesalski, H.K.; Chichili, G.R.; Frank, J.; von Lintig, J.; Nohr, D. Conversion of β-carotene to retinal pigment. Vitam. Horm. 2007, 75, 117–130. [Google Scholar] [PubMed]
- Feltl, L.; Pacakova, V.; Stulik, K.; Volka, K. Reliability of Carotenoid Analyses: A Review. Curr. Anal. Chem. 2005, 1, 93–102. [Google Scholar] [CrossRef]
- März, U. The Global Market for Carotenoids. Available online: https://www.bccresearch.com/market-Research/food-and-Beverage/carotenoids-Global-Market-Report-fod025e.html (accessed on 13 June 2017).
- Panis, G.; Carreon, J.R. Commercial astaxanthin production derived by green alga Haematococcus pluvialis: A microalgae process model and a techno-economic assessment all through production line. Algal Res. 2016, 18, 175–190. [Google Scholar] [CrossRef]
- Schweiggert, R.M.; Carle, R. Carotenoid Production by Bacteria, Microalgae, and Fungi. In Carotenoids Nutrition Analysis Technology; Kaczor, A., Baranska, M., Eds.; John Wiley & Sons, Ltd: Chichester, UK, 2016; pp. 217–240. [Google Scholar]
- Phaff, H.; Miller, M.; Yoneyama, M.; Soneda, M. A comparative study of the yeast florae associated with trees on the Japanese Islands and on the west coast of North America. In Proceedings of the 4th Internatoinal Fermentation Symposium: Fermentation Technology Today, Kyoto, Japan, 19–25 March 1972; pp. 759–774. [Google Scholar]
- Andrewes, A.G.; Starr, M.P. (3R,3′R)-Astaxanthin from the yeast Phaffia rhodozyma. Phytochemistry 1976, 15, 1009–1011. [Google Scholar] [CrossRef]
- Sandmann, G. Carotenoids of Biotechnological Importance. In Biotechnology of Isoprenoids; Schrader, J., Bohlmann, J., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 449–467. [Google Scholar] [CrossRef]
- Sharma, R.; Gassel, S.; Steiger, S.; Xia, X.; Bauer, R.; Sandmann, G.; Thines, M. The genome of the basal agaricomycete Xanthophyllomyces dendrorhous provides insights into the organization of its acetyl-CoA derived pathways and the evolution of Agaricomycotina. BMC Genom. 2015, 16, 233. [Google Scholar] [CrossRef] [PubMed]
- Visser, H.; Sandmann, G.; Verdoes, J.C. Xanthophylls in Fungi. Metabolic Engineering of the Astaxanthin Biosynthetic Pathway in Xanthophyllomyces dendrorhous. In Microbial Processes Products; Barredo, J.-L., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 257–272. [Google Scholar]
- Rodríguez-Sáiz, M.; Godio, R.P.; Alvarez, V.; de la Fuente, J.L.; Martín, J.F.; Barredo, J.L. The NADP-dependent glutamate dehydrogenase gene from the astaxanthin producer Xanthophyllomyces dendrorhous: Use of Its promoter for controlled gene expression. Mol. Biotechnol. 2009, 41, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Gassel, S.; Breitenbach, J.; Sandmann, G. Genetic engineering of the complete carotenoid pathway towards enhanced astaxanthin formation in Xanthophyllomyces dendrorhous starting from a high-yield mutant. Appl. Microbiol. Biotechnol. 2014, 98, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.Y.; Morita, T.; Mochizuki, M.; Yamamoto, K.; Ogino, C.; Araki, M.; Kondo, A. Development of a multi-gene expression system in Xanthophyllomyces dendrorhous. Microb. Cell Fact. 2014, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Yamane, Y.; Higashida, K.; Nakashimada, Y.; Kakizono, T.; Nishio, N. Influence of Oxygen and Glucose on Primary Metabolism and Astaxanthin Production by Phaffia rhodozyma in Batch and Fed-Batch Cultures: Kinetic and Stoichiometric Analysis. Appl. Environ. Microbiol. 1997, 63, 4471–4478. [Google Scholar] [PubMed]
- Alvarez, V.; Rodríguez-Sáiz, M.; de la Fuente, J.L.; Gudiña, E.J.; Godio, R.P.; Martín, J.F.; Barredo, J.L. The crtS gene of Xanthophyllomyces dendrorhous encodes a novel cytochrome-P450 hydroxylase involved in the conversion of β-carotene into astaxanthin and other xanthophylls. Fungal Genet. Biol. 2006, 43, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Ojima, K.; Breitenbach, J.; Visser, H.; Setoguchi, Y.; Tabata, K.; Hoshino, T.; van den Berg, J.; Sandmann, G. Cloning of the astaxanthin synthase gene from Xanthophyllomyces dendrorhous (Phaffia rhodozyma) and its assignment as a β-carotene 3-hydroxylase/4-ketolase. Mol. Genet. Genom. 2006, 275, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Green, A.S.; Fascetti, A.J. Meeting the Vitamin A Requirement: The Efficacy and Importance of β-Carotene in Animal Species. Sci. World J. 2016, 2016, 7393620. [Google Scholar] [CrossRef] [PubMed]
- Chichili, G.R.; Nohr, D.; Schäffer, M.; von Lintig, J.; Biesalski, H.K. β-Carotene conversion into vitamin A in human retinal pigment epithelial cells. Investig. Ophthalmol. Vis. Sci. 2005, 46, 3562–3569. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, K.Y.; Ding, X.M.; Bai, S.P.; Hernandez, J.M.; Yao, B.; Zhu, Q. Influence of canthaxanthin on broiler breeder reproduction, chick quality, and performance. Poult. Sci. 2011, 90, 1516–1522. [Google Scholar] [CrossRef] [PubMed]
- Bone, R.A.; Landrum, J.T.; Friedes, L.M.; Gomez, C.M.; Kilburn, M.D.; Menendez, E.; Vidal, I.; Wang, W. Distribution of lutein and zeaxanthin stereoisomers in the human retina. Exp. Eye Res. 1997, 64, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Landrum, J.T.; Bone, R.A. Lutein, zeaxanthin, and the macular pigment. Arch. Biochem. Biophys. 2001, 385, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Lima, V.C.; Rosen, R.B.; Farah, M. Macular pigment in retinal health and disease. Int. J. Retina Vitr. 2016, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Research and Markets, Global Astaxanthin Market-Sources, Technologies and Application. Available online: http://www.researchandmarkets.com/reports/3129287/global-astaxanthin-market-sources-technologies (accessed on 21 May 2017).
- Schmidt, I.; Schewe, H.; Gassel, S.; Jin, C.; Buckingham, J.; Hümbelin, M.; Sandmann, G.; Schrader, J. Biotechnological production of astaxanthin with Phaffia rhodozyma/Xanthophyllomyces dendrorhous. Appl. Microbiol. Biotechnol. 2011, 89, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Breithaupt, D.R. Xanthophylls in Poultry Feeding. In Carotenoids; Britton, G., Liaaen-Jensen, S., Pfander, H., Eds.; Birkhäuser Basel: Basel, Switzerland, 2008; Volume 4, pp. 255–264. [Google Scholar]
- Higuera-Ciapara, I.; Félix-Valenzuela, L.; Goycoolea, F.M. Astaxanthin: A review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006, 46, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Willén, R.; Wadström, T. Astaxanthin-rich algal meal and vitamin C inhibit Helicobacter pylori infection in BALB/cA mice. Antimicrob. Agents Chemother. 2000, 44, 2452–2457. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Chyun, J.H.; Kim, Y.K.; Line, L.L.; Chew, B.P. Astaxanthin decreased oxidative stress and inflammation and enhanced immune response in humans. Nutr. Metab. 2010, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Hosokawa, M.; Mikami, N.; Miyashita, K.; Tanaka, T. Dietary astaxanthin inhibits colitis and colitis-associated colon carcinogenesis in mice via modulation of the inflammatory cytokines. Chem. Biol. Interact. 2011, 193, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Fassett, R.G.; Coombes, J.S. Astaxanthin, oxidative stress, inflammation and cardiovascular disease. Future Cardiol. 2009, 5, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, D.; Niu, J.; Shen, S.; Wang, G. An economic assessment of astaxanthin production by large scale cultivation of Haematococcus pluvialis. Biotechnol. Adv. 2011, 29, 568–574. [Google Scholar] [CrossRef] [PubMed]
- An, G.H.; Cho, M.H.; Johnson, E.A. Monocyclic carotenoid biosynthetic pathway in the yeast Phaffia rhodozyma (Xanthophyllomyces dendrorhous). J. Biosci. Bioeng. 1999, 88, 189–193. [Google Scholar] [CrossRef]
- Rodríguez-Sáiz, M.; de la Fuente, J.L.; Barredo, J.L. Xanthophyllomyces dendrorhous for the industrial production of astaxanthin. Appl. Microbiol. Biotechnol. 2010, 88, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Priya, R.; Hridya, H.; Soundarya, C.; Somasundari, G.; Doss, C.G.P.; Sneha, P.; Rajasekaran, C.; Christopher, G.; Siva, R. Astaxanthin biosynthetic pathway: Molecular phylogenies and evolutionary behaviour of Crt genes in eubacteria. Plant Gene 2016, 8, 32–41. [Google Scholar] [CrossRef]
- Andrewes, A.G.; Phaff, H.J.; Starr, M.P. Carotenoids of Phaffia rhodozyma, a red-pigmented fermenting yeast. Phytochemistry 1976, 15, 1003–1007. [Google Scholar] [CrossRef]
- Sanpietro, L.M.D.; Kula, M.R. Studies of astaxanthin biosynthesis in Xanthophyllomyces dendrorhous (Phaffia rhodozyma). Effect of inhibitors and low temperature. Yeast 1998, 14, 1007–1016. [Google Scholar] [CrossRef]
- Verdoes, J.C.; Sandmann, G.; Visser, H.; Diaz, M.; van Mossel, M.; van Ooyen, A.J.J. Metabolic engineering of the carotenoid biosynthetic pathway in the yeast Xanthophyllomyces dendrorhous (Phaffia rhodozyma). Appl. Environ. Microbiol. 2003, 69, 3728–3738. [Google Scholar] [CrossRef] [PubMed]
- Visser, H.; van Ooyen, A.J.J.; Verdoes, J.C. Metabolic engineering of the astaxanthin-biosynthetic pathway of Xanthophyllomyces dendrorhous. FEMS Yeast Res. 2003, 4, 221–231. [Google Scholar] [CrossRef]
- Liang, P.-H.; Ko, T.-P.; Wang, A.H.-J. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002, 269, 3339–3354. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Schmidt-Dannert, C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl. Microbiol. Biotechnol. 2002, 60, 1–11. [Google Scholar] [PubMed]
- Misawa, N. Pathway engineering for functional isoprenoids. Curr. Opin. Biotechnol. 2011, 22, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, S.; Fraser, P.D.; Kondo, K.; Misawa, N. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 1997, 324, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Britton, G. Carotenoid Biosynthesis—An Overview. In Carotenoids Chemistry Biology; Krinsky, N.I., Mathews-Roth, M.M., Taylor, R.F., Eds.; Springer: Boston, MA, USA, 1989; pp. 167–184. [Google Scholar]
- Niklitschek, M.; Alcaíno, J.; Barahona, S.; Sepúlveda, D.; Lozano, C.; Carmona, M.; Marcoleta, A.; Martínez, C.; Lodato, P.; Baeza, M.; et al. Genomic organization of the structural genes controlling the astaxanthin biosynthesis pathway of Xanthophyllomyces dendrorhous. Biol. Res. 2008, 41, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Verdoes, J.C.; Krubasik, K.P.; Sandmann, G.; van Ooyen, A.J. Isolation and functional characterisation of a novel type of carotenoid biosynthetic gene from Xanthophyllomyces dendrorhous. Mol. Gen. Genet. 1999, 262, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Ajikumar, P.K.; Tyo, K.; Carlsen, S.; Mucha, O.; Phon, T.H.; Stephanopoulos, G. Terpenoids: Opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol. Pharm. 2008, 5, 167–190. [Google Scholar] [CrossRef] [PubMed]
- Verdoes, J.C.; Misawa, N.; van Ooyen, A.J. Cloning and characterization of the astaxanthin biosynthetic gene encoding phytoene desaturase of Xanthophyllomyces dendrorhous. Biotechnol. Bioeng. 1999, 63, 750–755. [Google Scholar] [CrossRef]
- Krubasik, P.; Sandmann, G. A carotenogenic gene cluster from Brevibacterium linens with novel lycopene cyclase genes involved in the synthesis of aromatic carotenoids. Mol. Gen. Genet. 2000, 263, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Alcaíno, J.; Barahona, S.; Carmona, M.; Lozano, C.; Marcoleta, A.; Niklitschek, M.; Sepúlveda, D.; Baeza, M.; Cifuentes, V. Cloning of the cytochrome p450 reductase (crtR) gene and its involvement in the astaxanthin biosynthesis of Xanthophyllomyces dendrorhous. BMC Microbiol. 2008, 8, 169. [Google Scholar] [CrossRef] [PubMed]
- Ukibe, K.; Hashida, K.; Yoshida, N.; Takagi, H. Metabolic engineering of Saccharomyces cerevisiae for astaxanthin production and oxidative stress tolerance. Appl. Environ. Microbiol. 2009, 75, 7205–7211. [Google Scholar] [CrossRef] [PubMed]
- Calo, P.; González, T. The yeast Phaffia rhodozyma as an industrial source of astaxanthin. Microbiologia 1995, 11, 386–388. [Google Scholar] [PubMed]
- Alcaino, J.; Baeza, M.; Cifuentes, V. Astaxanthin and Related Xanthophylls. In Biosynthesis Molecular Genetics of Fungal Secondary Metabolites; Martín, J.-F., Garcia-Estrada, C., Zeilinger, S., Eds.; Springer: New York, NY, USA, 2014; pp. 187–207. [Google Scholar]
- Ukibe, K.; Katsuragi, T.; Tani, Y.; Takagi, H. Efficient screening for astaxanthin-overproducing mutants of the yeast Xanthophyllomyces dendrorhous by flow cytometry. FEMS Microbiol. Lett. 2008, 286, 241–248. [Google Scholar] [CrossRef] [PubMed]
- An, G.H.; Schuman, D.B.; Johnson, E.A. Isolation of Phaffia rhodozyma Mutants with Increased Astaxanthin Content. Appl. Environ. Microbiol. 1989, 55, 116–124. [Google Scholar] [PubMed]
- Retamales, P.; León, R.; Martínez, C.; Hermosilla, G.; Pincheira, G.; Cifuentes, V. Complementation analysis with new genetic markers in Phaffia rhodozyma. Antonie Van Leeuwenhoek 1998, 73, 229–236. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente Moreno, J.L.; Peiro, E.; Díez García, B.; Marcos Rodríguez, A.T.; Schleissner, C.; Rodríguez Saiz, M.; Rodríguez-Otero, C.; Cabri, W.; Barredo, J.L. Method of Producing Astaxanthin by Fermenting Selected Strains of Xanthophyllomyces dendrorhous. Patent EP1479777A1, 3 February 2003. [Google Scholar]
- Liu, Z.Q.; Zhang, J.F.; Zheng, Y.G.; Shen, Y.C. Improvement of astaxanthin production by a newly isolated Phaffia rhodozyma mutant with low-energy ion beam implantation. J. Appl. Microbiol. 2008, 104, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Hong, Q.; Xiao, A.; Li, L.; Cai, H.; Su, W. Characterization and evaluation of an astaxanthin over-producing Phaffia rhodozyma. Sheng Wu Gong Cheng Xue Bao (Chinese J. Biotechnol.) 2011, 27, 1065–1075. [Google Scholar]
- Johnson, E.A. Phaffia rhodozyma: Colorful odyssey. Int. Microbiol. 2003, 6, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Chumpolkulwong, N.; Kakizono, T.; Nagai, S.; Nishio, N. Increased astaxanthin production by Phaffia rhodozyma mutants isolated as resistant to diphenylamine. J. Ferment. Bioeng. 1997, 83, 429–434. [Google Scholar] [CrossRef]
- Schroeder, W.A.; Calo, P.; DeClercq, M.L.; Johnson, E.A. Selection for carotenogenesis in the yeast Phaffia rhodozyma by dark-generated singlet oxygen. Microbiology 1996, 142, 2923–2929. [Google Scholar] [CrossRef]
- An, G.H.; Johnson, E.A. Influence of light on growth and pigmentation of the yeast Phaffia rhodozyma. Antonie Van Leeuwenhoek 1990, 57, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, F.; Vasco-Cárdenas, M.F.; Barreiro, C. Biotypes analysis of Corynebacterium glutamicum growing in dicarboxylic acids demonstrates the existence of industrially-relevant intra-species variations. J. Proteom. 2016, 146, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Breitenbach, J.; Visser, H.; Verdoes, J.C.; van Ooyen, A.J.J.; Sandmann, G. Engineering of geranylgeranyl pyrophosphate synthase levels and physiological conditions for enhanced carotenoid and astaxanthin synthesis in Xanthophyllomyces dendrorhous. Biotechnol. Lett. 2011, 33, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Gassel, S.; Schewe, H.; Schmidt, I.; Schrader, J.; Sandmann, G. Multiple improvement of astaxanthin biosynthesis in Xanthophyllomyces dendrorhous by a combination of conventional mutagenesis and metabolic pathway engineering. Biotechnol. Lett. 2013, 35, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Wery, J.; Verdoes, J.C.; van Ooyen, A.J.J. Efficient Transformation of the Astaxanthin-Producing Yeast Phaffia rhodozyma. Biotechnol. Tech. 1998, 12, 399–405. [Google Scholar] [CrossRef]
- Yamamoto, K.; Hara, K.Y.; Morita, T.; Nishimura, A.; Sasaki, D.; Ishii, J.; Ogino, C.; Kizaki, N.; Kondo, A. Enhancement of astaxanthin production in Xanthophyllomyces dendrorhous by efficient method for the complete deletion of genes. Microb. Cell Fact. 2016, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.Y.; Morita, T.; Endo, Y.; Mochizuki, M.; Araki, M.; Kondo, A. Evaluation and screening of efficient promoters to improve astaxanthin production in Xanthophyllomyces dendrorhous. Appl. Microbiol. Biotechnol. 2014, 98, 6787–6793. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Gudiña, E.; Barredo, J.L. Conversion of β-carotene into astaxanthin: Two separate enzymes or a bifunctional hydroxylase-ketolase protein? Microb. Cell Fact. 2008, 7, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodato, P.; Alcaíno, J.; Barahona, S.; Niklitschek, M.; Carmona, M.; Wozniak, A.; Baeza, M.; Jiménez, A.; Cifuentes, V. Expression of the carotenoid biosynthesis genes in Xanthophyllomyces dendrorhous. Biol. Res. 2007, 40, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Chi, S.; He, Y.; Ren, J.; Su, Q.; Liu, X.; Chen, Z.; Wang, M.; Li, Y.; Li, J. Overexpression of a bifunctional enzyme, CrtS, enhances astaxanthin synthesis through two pathways in Phaffia rhodozyma. Microb. Cell Fact. 2015, 14, 90. [Google Scholar] [CrossRef] [PubMed]
- Ledetzky, N.; Osawa, A.; Iki, K.; Pollmann, H.; Gassel, S.; Breitenbach, J.; Shindo, K.; Sandmann, G. Multiple transformation with the crtYB gene of the limiting enzyme increased carotenoid synthesis and generated novel derivatives in Xanthophyllomyces dendrorhous. Arch. Biochem. Biophys. 2014, 545, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.G.; Hu, Z.C.; Wang, Z.; Shen, Y.C. Large-Scale Production of Astaxanthin by Xanthophyllomyces dendrorhous. Food Bioprod. Process. 2006, 84, 164–166. [Google Scholar] [CrossRef]
- De la Fuente, J.L.; Rodríguez-Sáiz, M.; Schleissner, C.; Díez, B.; Peiro, E.; Barredo, J.L. High-titer production of astaxanthin by the semi-industrial fermentation of Xanthophyllomyces dendrorhous. J. Biotechnol. 2010, 148, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, V.; Hermosilla, G.; Martínez, C.; León, R.; Pincheira, G.; Jiménez, A. Genetics and electrophoretic karyotyping of wild-type and astaxanthin mutant strains of Phaffia rhodozyma. Antonie Van Leeuwenhoek 1997, 72, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Baeza, M.; Alcaíno, J.; Barahona, S.; Sepúlveda, D.; Cifuentes, V. Codon usage and codon context bias in Xanthophyllomyces dendrorhous. BMC Genom. 2015, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.; Lozano, C.; Barahona, S.; Niklitschek, M.; Marcoleta, A.; Alcaíno, J.; Sepulveda, D.; Baeza, M.; Cifuentes, V. Differential carotenoid production and gene expression in Xanthophyllomyces dendrorhous grown in a nonfermentable carbon source. FEMS Yeast Res. 2011, 11, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Bellora, N.; Moliné, M.; David-Palma, M.; Coelho, M.A.; Hittinger, C.T.; Sampaio, J.P.; Gonçalves, P.; Libkind, D. Comparative genomics provides new insights into the diversity, physiology, and sexuality of the only industrially exploited tremellomycete: Phaffia rhodozyma. BMC Genom. 2016, 17, 901. [Google Scholar] [CrossRef] [PubMed]
- Verdoes, J.C.; van Ooyen, A.J.J. Codon usage in Xanthophyllomyces dendrorhous (formerly Phaffia rhodozyma). Biotechnol. Lett. 2000, 22, 9–13. [Google Scholar] [CrossRef]
- Elena, C.; Ravasi, P.; Castelli, M.E.; Peirú, S.; Menzella, H.G. Expression of codon optimized genes in microbial systems: Current industrial applications and perspectives. Front. Microbiol. 2014, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Barbachano-Torres, A.; Castelblanco-Matiz, L.M.; Ramos-Valdivia, A.C.; Cerda-García-Rojas, C.M.; Salgado, L.M.; Flores-Ortiz, C.M.; Ponce-Noyola, T. Analysis of proteomic changes in colored mutants of Xanthophyllomyces dendrorhous (Phaffia rhodozyma). Arch. Microbiol. 2014, 196, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Moya, P.; Niehaus, K.; Alcaíno, J.; Baeza, M.; Cifuentes, V. Proteomic and metabolomic analysis of the carotenogenic yeast Xanthophyllomyces dendrorhous using different carbon sources. BMC Genom. 2015, 16, 289. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Moya, P.; Watt, S.A.; Niehaus, K.; Alcaíno, J.; Baeza, M.; Cifuentes, V. Proteomic analysis of the carotenogenic yeast Xanthophyllomyces dendrorhous. BMC Microbiol. 2011, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Wang, B.; Gerken, H.G.; Lu, Y.; Ling, X. Proteomic analysis of astaxanthin biosynthesis in Xanthophyllomyces dendrorhous in response to low carbon levels. Bioprocess Biosyst. Eng. 2017. [CrossRef] [PubMed]
- Barreiro, C.; Martín, J.F.; García-Estrada, C. Proteomics Shows New Faces for the Old Penicillin Producer Penicillium chrysogenum. J. Biomed. Biotechnol. 2012, 2012, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.; Brent, R.; Kingston, R.; Moore, D.; Seidman, J.; Smith, J.; Struhl, K. YPD media. In Current Protocols Molecular Biology Cold Spring Harbor Protocols; Cold Spring Harbor Laboratory Press: Brooklyn, NY, USA, 1994. [Google Scholar] [CrossRef]
- Gonçalves, P.; Valério, E.; Correia, C.; de Almeida, J.M.G.C.F.; Sampaio, J.P. Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLoS ONE 2011, 6, e20739. [Google Scholar] [CrossRef] [PubMed]
- Soni, R.; Murray, J.A. A rapid and inexpensive method for isolation of shuttle vector DNA from yeast for the transformation of E. coli. Nucleic Acids Res. 1992, 20, 5852. [Google Scholar] [CrossRef] [PubMed]
- Retamales, P.; Hermosilla, G.; León, R.; Martínez, C.; Jiménez, A.; Cifuentes, V. Development of the sexual reproductive cycle of Xanthophyllomyces dendrorhous. J. Microbiol. Methods 2002, 48, 87–93. [Google Scholar] [CrossRef]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef] [PubMed]

| Targets | Approach | Result | Ref. |
|---|---|---|---|
| crtYB gene | Deactivation | No carotenoids | [48] |
| crtYB gene | Overexpression | Accumulation of β-carotene and echinenone | [48] |
| crtI gene | Overexpression | Increase torulene and HDCO and decrease echinenone, l-carotene and astaxanthin | [48] |
| crtR gene | Description of its role | Required together with the crtS gene for the conversion of β-carotene to astaxanthin. | [59] |
| double cyp61 genes | Deletion | Enhanced astaxanthin production by 1,4-fold compared with the parental strain | [77] |
| crtE gene under Padh4r | Evaluation of promotors | Increase in intracellular astaxanthin by 1.7-fold compared with parental | [78] |
| acaT, hmgS and hmgR genes | Triple overexpression | Enhanced volumetric astaxanthin production by 1.4-fold compared with that of the control strain | [23] |
| acaT/hmgS/hmgR/crtE/crtS genes | Combined overexpression | Enhanced volumetric astaxanthin production by 2.1-fold higher compared with the control strain | [23] |
| Combination of conventional mutagenesis and crtYB gene expression | Combined overexpression | 22 times higher astaxanthin specific production than for the wild type | [75] |
| Media | Collection | Disruption | Analysis Method | Ref. |
|---|---|---|---|---|
| DNA | ||||
| YPD medium. 21 °C, 5 days [96] | Culture: 15 mL Suspended: 0.5 mL YPD | Breaking system: 300 μL of glass beads (0.25–0.5 mm diameter). Swing mill (Retsch MM200) at a frequency of 30/s. | Agarose gel and ethidium bromide stain. Fluorescence densitometry measurement. | [19] |
| Sample cleaning: Supernatant collected and purified by phenol/chloroform/isoamyl alcohol extraction. DNA precipitation overnight at −20 °C by 100% ice-cold ethanol (2.5 volume) and 1/10 volume of 3 M sodium acetate solution. 70% ice-cold ethanol. Dry at room temperature. DNA pellet resuspension in 30 μL H2O. Store at 4 °C. | ||||
| YM medium (100 mL). 22 °C, up to stationary phase | Centrifugation | Breaking system: DNA isolated from protoplasts: 2× wash 50 mM EDTA pH 7.5. Novozyme 234 plus LET buffer (500 mM EDTA, 7.5% 2-mercaptoethanol, 10 mM Tris pH 7.5). 16 h, 37 °C. NDS solution. (2 mg/mL proteinase K in 500 mM EDTA, 1% lauryl sarcosine and 10 mM Tris-HCl, pH 7.5). 24 h, 50 °C. | DNA quantitation: 260/280 ratio (1.7–1.9) and 260/230 ratio (>2) by using a V-630 UV–vis Spectrophotometer. | [85,86] |
| Sample cleaning: Phenolic extraction (pH 8.0): 3× wash saturated phenol. 3× phenol: chloroform: isoamyl alcohol (25:24:1). 1× chloroform: isoamyl alcohol (24:1). DNA was precipitated with 98% ethanol and washed with 70% ethanol. Dry DNA resuspended in Tris: EDTA (10:1; pH 8.0) plus 40 μg/mL of RNase A. 37 °C, 30 min. Repeat phenolic extraction | ||||
| YM broth (15 mL) at 20 °C, 72 h | Centrifugation | Breaking system: Modified phenol:chloroform:isoamyl alcohol method [97,98]: Resuspend in 500 µL lysis buffer (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA (pH 8.0), 2% Triton X-100, 1% SDS). Add an equal volume of phenol/chloroform (1∶1 v/v). Shake vigorously (Ika-Vibrax VXR shaker) at 1800 rpm, 20 minutes, R/T. Centrifuge at 14,000 rpm, 20 min, 4 °C. | N/A | [88] |
| Sample cleaning: Ethanol precipitation. | ||||
| RNA | ||||
| YPD medium. 21 °C, 5 days [96] | NucleoSpin® RNA Plant kit (MACHEREY-NAGEL GmbH & Co. KG) (Following the manufacturer instructions). | RNA quality by using Nano-Photometer (IMPLEN) 1.5% agarose gel and ethidium bromide stain | [19] | |
| Vogel minimal medium (MMv) supplemented with 2% glucose or 2% succinate | Early exponential phase (18 h) Initial stationary phase (72 h) | Breaking system: Mechanical rupture of cell pellets. 0.5 mm glass beads (BioSpec). Vortexing for 10 min. Add Tri-Reagent (Ambion). R/T 10 min. | RNA quantitation: 260/280 ratio (>1.9) by using a V-630 UV–vis Spectrophotometer | [86] |
| Sample cleaning: Add 200 μL of chloroform per mL of Tri-Reagent. Mix. Centrifuge: 4000× g, 5 min. Recover supernatant. 2× acidic phenol: chloroform (1:1) extractions. Precipitate: 2 volume of isopropanol. 10 min, R/T. 1× wash 75% ethanol. Resuspend in RNase-free water. RNA samples at a 260/280 ratio >1.9, measured using a V-630 UV–vis Spectrophotometer, were used for next-generation sequencing. | ||||
| Proteins | ||||
| Minimal medium plus 2% glucose or succinate as carbon sources [99] Preculture: 10 mL Culture: 250 mL in 1-L flask inoculated with 2.5 mL of seed culture. 22 °C, 120 rpm | Centrifugation: 5000× g, 10 min, 4 °C. Pellet washed twice with ice-cold water. Centrifuge: 5000 × g, 10 min, 4 °C. Freeze in liquid N2. Stored at −80 °C | Breaking system: Lyophilise cells. Add an equal volume (±500 μL) of glass beads (500 μm). Add 500 μL of lysis buffer (100 mM sodium. bicarbonate, pH 8.8, 0.5% Triton × 100, 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors). 15 min in on ice. Shake at 30 s at 4.5 m/s (RiboLyzer). Chill on ice, 1 min between shaking steps. | Bidimensional gel (pI range: 3–10 NL, 17 cm strips) Coomassie brilliant blue Trypsin digestion MALDI-TOF-MS identification | [92,93] |
| Sample cleaning: Remove cell debris by centrifugation (15,000 rpm, 20 min, 4 °C. 10% v/v DNase-RNase solution (0.5 M Tris-HCl, pH 7.0, 0.5 M MgCl2, 100 μg/mL RNAse and 2 μL DNase). 1 h, 4 °C. Add water up to 2.5 mL plus 200 μL of 0.5 M Tris (pH 6.8) and 20 μL of 1 M dithiothreitol (DTT). R/T. 30 min. 600 μL of water-saturated phenol. R/T. 30 min. Centrifuge: 5000× g, 10 min, 4 °C Add to supernatant 20 μL of 1 M DTT and 30 μL of 8 M ammonium acetate. R/T. 30 min. Precipitate. 2 mL of cold (−20 °C) methanol. Centrifuge: 13,000 rpm, 4 °C, 30 min. 2× wash: 70% (v/v) cold ethanol at −20 °C. Resuspend pellet: 200 μL of buffer (8 M urea, 2 M thiourea, 2% CHAPS, 0.01% [w/v] bromophenol blue). Store at −80 °C. | ||||
| YM medium 20-h old culture (beginning of carotenoid biosynthesis) 20 °C and 150 rpm. | Centrifugation: 5000 rpm, 10 min. | Breaking system: Liquid nitrogen in a mortar Resuspend fine powder in 5 mL of buffer (50 mM Tris–HCl pH 7.4, 0.5 mM PMSF). Add 5 mL of 100 mM sodium carbonate. 1 h on ice. Centrifuge: 6000 rpm, 20 min, 4 °C. Supernatant precipitation: 10% final concentration of trichloroacetic acid (TCA). Centrifuge: 10,000 rpm for 15 min. | Bidimensional gel (pI range: 3–10, 11 cm strips) Colloidal Coomassie [100] | [91] |
| Sample cleaning: 1× acetone. 1× 70% ethanol. 200 μL rehydration buffer (7 M urea, 2 M thiourea, 1% CHAPS, 0.5% Triton X-100, 40 mM Tris–HCl, 0.5% ampholytes 3–10, and 0.1% bromophenol blue). Centrifuge: 10,000 rpm, 15 min. Desalt: Micro Bio Spin G-30 columns (Bio-Rad) and rehydration buffer (Bio-Rad, Hercules, CA, USA). | ||||
| Seed culture: 20 g/L glucose, 10 g/L yeast extract, 20 g/L peptone. 22 °C, 200 rpm, 48 h. 250-mL flask containing 30 mL Inoculate 3 mL in a 250-mL flask containing 30 mL of seed media. 22 °C, 200 rpm, 24 h Fermentation: (20 g/L glucose, 0.2 g/L yeast extract, 0.5 g/L (NH4)2SO4, 1.0 g/L KH2PO4, 0.1 g/L NaCl, 0.5 g/L MgSO4-7H2O, and 0.1 g/L CaCl2-2H2O). 22 °C, 200 rpm, 144 h | Centrifugation: 8000× g, 10 min, 8 °C, 96 h. | Breaking system: Resuspend pellet in deionised water. 2× 30 kpsi high-pressure disrupter (Constant Systems Limited, Northants, UK). 10,000× g, 15 min, 4 °C. Collect supernatant 0.5 mg dissolved in 500 µL rehydration buffer [8 M urea, 4% CHAPS, 2% IPG buffer and 40 mM DTT (Dithiothreitol)]. | Bidimensional gel (pI range: 4–7) Silver stain Trypsin digestion MALDI-TOF/TOF-MS identification | [94] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barredo, J.L.; García-Estrada, C.; Kosalkova, K.; Barreiro, C. Biosynthesis of Astaxanthin as a Main Carotenoid in the Heterobasidiomycetous Yeast Xanthophyllomyces dendrorhous. J. Fungi 2017, 3, 44. https://doi.org/10.3390/jof3030044
Barredo JL, García-Estrada C, Kosalkova K, Barreiro C. Biosynthesis of Astaxanthin as a Main Carotenoid in the Heterobasidiomycetous Yeast Xanthophyllomyces dendrorhous. Journal of Fungi. 2017; 3(3):44. https://doi.org/10.3390/jof3030044
Chicago/Turabian StyleBarredo, Jose L., Carlos García-Estrada, Katarina Kosalkova, and Carlos Barreiro. 2017. "Biosynthesis of Astaxanthin as a Main Carotenoid in the Heterobasidiomycetous Yeast Xanthophyllomyces dendrorhous" Journal of Fungi 3, no. 3: 44. https://doi.org/10.3390/jof3030044