Conserved Inhibition of Neutrophil Extracellular Trap Release by Clinical Candida albicans Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Inoculum
2.2. Microfluidic Device Model and Imaging
2.3. Biofilm Formation
2.4. Human Neutrophil Collection
2.5. Scanning Electron Microscopy
2.6. Measurement of ROS
2.7. Sytox Green Assays
2.8. Fluorescent Imaging
2.9. Statistics
3. Results
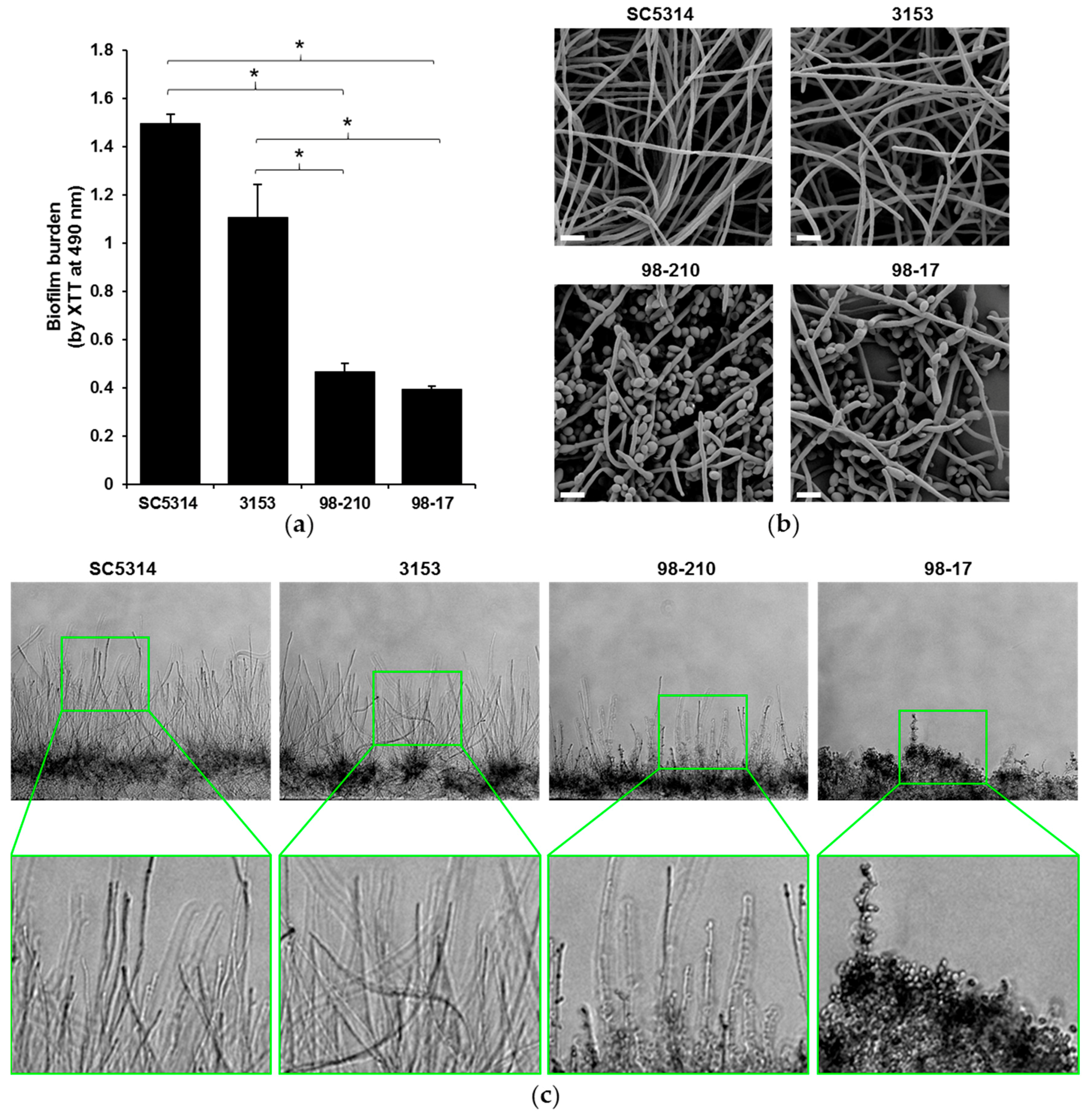
3.1. Biofilms Produced by C. albicans Clinical Isolates Differ in Architecture and Thickness
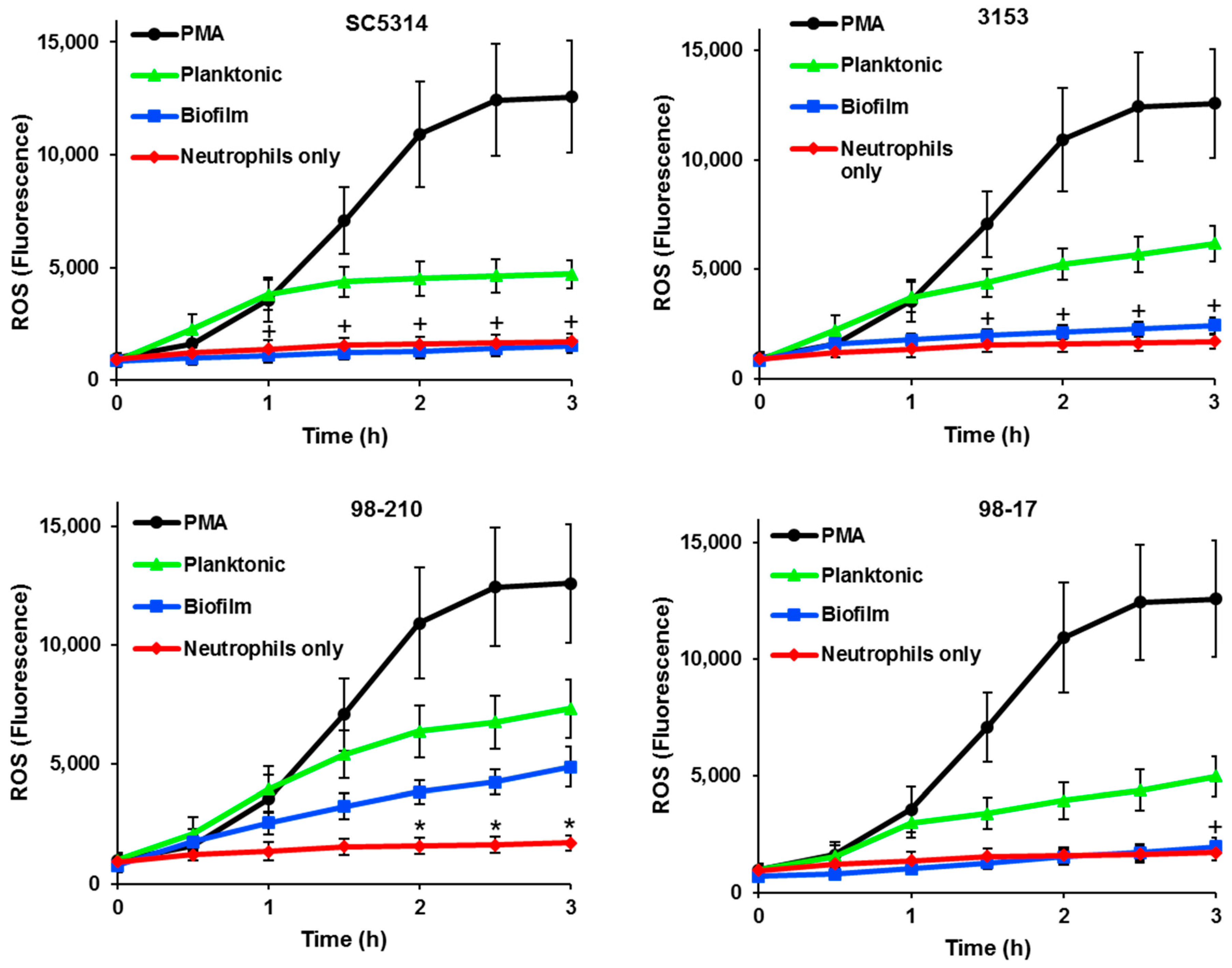
3.2. C. albicans Biofilms Variably Inhibit ROS Production by Neutrophils
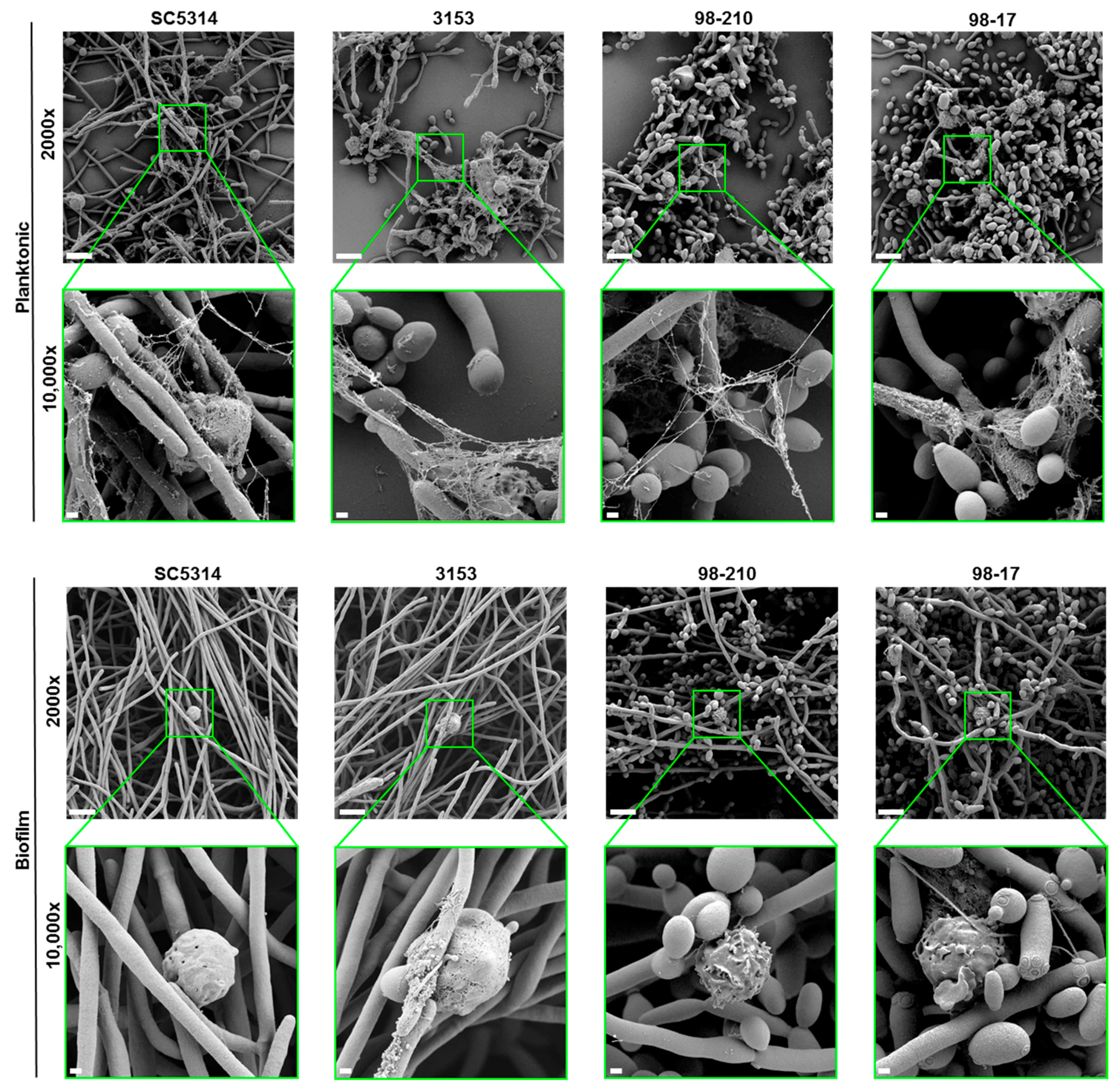
3.3. C. albicans Biofilms Uniformly Inhibit the Formation of NETs
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Dongari-Bagtzoglou, A. Mucosal biofilms: Challenges and future directions. Expert Rev. Anti Infect. Ther. 2008, 6, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Kojic, E.M.; Darouiche, R.O. Candida infections of medical devices. Clin. Microbiol. Rev. 2004, 17, 255–267. [Google Scholar] [PubMed]
- Kumamoto, C.A.; Vinces, M.D. Alternative Candida albicans lifestyles: Growth on surfaces. Annu. Rev. Microbiol. 2005, 59, 113–133. [Google Scholar] [CrossRef] [PubMed]
- Katragkou, A.; Kruhlak, M.J.; Simitsopoulou, M.; Chatzimoschou, A.; Taparkou, A.; Cotten, C.J.; Paliogianni, F.; Diza-Mataftsi, E.; Tsantali, C.; Walsh, T.J.; et al. Interactions between human phagocytes and Candida albicans biofilms alone and in combination with antifungal agents. J. Infect. Dis. 2010, 201, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Katragkou, A.; Simitsopoulou, M.; Chatzimoschou, A.; Georgiadou, E.; Walsh, T.J.; Roilides, E. Effects of interferon-gamma and granulocyte colony-stimulating factor on antifungal activity of human polymorphonuclear neutrophils against Candida albicans grown as biofilms or planktonic cells. Cytokine 2011, 55, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Chandra, J.; Kuhn, D.M.; Mukherjee, P.K.; Hoyer, L.L.; McCormick, T.; Ghannoum, M.A. Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. J. Bacteriol. 2001, 183, 5385–5394. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Bachmann, S.; Patterson, T.F.; Wickes, B.L.; Lopez-Ribot, J.L. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 2002, 49, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Rajendran, R.; Sherry, L.; Williams, C. Fungal biofilm resistance. Int. J. Microbiol. 2012, 2012, 528521. [Google Scholar] [PubMed]
- Shoham, S.; Levitz, S.M. The immune response to fungal infections. Br. J. Haematol. 2005, 129, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.E., Jr.; Lehrer, R.I.; Stiehm, E.R.; Fischer, T.J.; Young, L.S. Severe candidal infections: Clinical perspective, immune defense mechanisms, and current concepts of therapy. Ann. Intern. Med. 1978, 89, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Fidel, P.L., Jr. Immunity to Candida. Oral Dis. 2002, 8 (Suppl. 2), 69–75. [Google Scholar] [CrossRef] [PubMed]
- Erwig, L.P.; Gow, N.A. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Alexander, B.; Brumble, L.; Freifeld, A.; Hadley, S.; Herwaldt, L.; Kauffman, C.; Lyon, G.M.; et al. The epidemiology and outcomes of invasive Candida infections among organ transplant recipients in the United States: Results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Transpl. Infect. Dis. 2016, 18, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.R.; Safdar, N.; Baddley, J.W.; Playford, G.; Reboli, A.C.; Rex, J.H.; Sobel, J.D.; Pappas, P.G.; Kullberg, B.J. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: A patient-level quantitative review of randomized trials. Clin. Infect. Dis. 2012, 54, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Thompson, A.; Sobue, T.; Kashleva, H.; Xu, H.; Vasilakos, J.; Dongari-Bagtzoglou, A. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J. Infect. Dis. 2012, 206, 1936–1945. [Google Scholar] [CrossRef] [PubMed]
- Katragkou, A.; Chatzimoschou, A.; Simitsopoulou, M.; Georgiadou, E.; Roilides, E. Additive antifungal activity of anidulafungin and human neutrophils against Candida parapsilosis biofilms. J. Antimicrob. Chemother. 2011, 66, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.J.; Cabezas-Olcoz, J.; Kernien, J.F.; Wang, S.X.; Beebe, D.J.; Huttenlocher, A.; Ansari, H.; Nett, J.E. The extracellular matrix of Candida albicans biofilms impairs formation of neutrophil extracellular traps. PLoS Pathog. 2016, 12, e1005884. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.F.; Reichard, U.; Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 2006, 8, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.S.; O’Brien, X.M.; Laforce-Nesbitt, S.S.; Parisi, V.E.; Hirakawa, M.P.; Bliss, J.M.; Reichner, J.S. NETosis in Neonates: Evidence of a Reactive Oxygen Species-Independent Pathway in Response to Fungal Challenge. J. Infect. Dis. 2016, 213, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Hopke, A.; Nicke, N.; Hidu, E.E.; Degani, G.; Popolo, L.; Wheeler, R.T. Neutrophil attack triggers extracellular trap-dependent Candida cell wall remodeling and altered immune recognition. PLoS Pathog. 2016, 12, e1005644. [Google Scholar] [CrossRef] [PubMed]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Wozniok, I.; Hornbach, A.; Schmitt, C.; Frosch, M.; Einsele, H.; Hube, B.; Loffler, J.; Kurzai, O. Induction of ERK-kinase signalling triggers morphotype-specific killing of Candida albicans filaments by human neutrophils. Cell. Microbiol. 2008, 10, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Gantner, B.N.; Simmons, R.M.; Underhill, D.M. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J. 2005, 24, 1277–1286. [Google Scholar] [CrossRef] [PubMed]
- Warnatsch, A.; Tsourouktsoglou, T.D.; Branzk, N.; Wang, Q.; Reincke, S.; Herbst, S.; Gutierrez, M.; Papayannopoulos, V. Reactive Oxygen Species Localization Programs Inflammation to Clear Microbes of Different Size. Immunity 2017, 46, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.; Diekema, D.J.; Pfaller, M.A.; Prince, R.A.; Marchillo, K.; Ashbeck, J.; Hou, J. In vivo pharmacodynamic characterization of anidulafungin in a neutropenic murine candidiasis model. Antimicrob. Agents Chemother. 2008, 52, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Romeo, O.; Criseo, G. First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene. Diagn. Microbiol. Infect. Dis. 2008, 62, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Cain, M.T.; Crawford, K.; Andes, D.R. Optimizing a Candida biofilm microtiter plate model for measurement of antifungal susceptibility by tetrazolium salt assay. J. Clin. Microbiol. 2011, 49, 1426–1433. [Google Scholar] [CrossRef] [PubMed]
- Taff, H.T.; Nett, J.E.; Andes, D.R. Comparative analysis of Candida biofilm quantitation assays. Med. Mycol. 2012, 50, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, R.; Leonelli, L.; Han, H.; Grigoryev, S.A.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E. The Host’s Reply to Candida Biofilm. Pathogens 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Kenny, E.F.; Herzig, A.; Kruger, R.; Muth, A.; Mondal, S.; Thompson, P.R.; Brinkmann, V.; Von Bernuth, H.; Zychlinsky, A. Diverse stimuli engage different neutrophil extracellular trap pathways. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, H.; Welin, A.; Michaelsson, E.; Osla, V.; Berg, S.; Christenson, K.; Sundqvist, M.; Dahlgren, C.; Karlsson, A.; Bylund, J. Neutrophil NET formation is regulated from the inside by myeloperoxidase-processed reactive oxygen species. Free Radic. Biol. Med. 2015, 89, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Stoiber, W.; Obermayer, A.; Steinbacher, P.; Krautgartner, W.D. The Role of Reactive Oxygen Species (ROS) in the Formation of Extracellular Traps (ETs) in Humans. Biomolecules 2015, 5, 702–723. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.D.; Lucas, C.D.; Mackellar, A.; Li, F.; Hiersemenzel, K.; Haslett, C.; Davidson, D.J.; Rossi, A.G. Activation of conventional protein kinase C (PKC) is critical in the generation of human neutrophil extracellular traps. J. Inflamm. 2013, 10, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, D.; Wheeler, R.T. The complex roles of NADPH oxidases in fungal infection. Cell. Microbiol. 2014, 16, 1156–1167. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.S.; O’Brien, X.M.; Johnson, C.M.; Lavigne, L.M.; Reichner, J.S. An extracellular matrix-based mechanism of rapid neutrophil extracellular trap formation in response to Candida albicans. J. Immunol. 2013, 190, 4136–4148. [Google Scholar] [CrossRef] [PubMed]
- Metzler, K.D.; Fuchs, T.A.; Nauseef, W.M.; Reumaux, D.; Roesler, J.; Schulze, I.; Wahn, V.; Papayannopoulos, V.; Zychlinsky, A. Myeloperoxidase is required for neutrophil extracellular trap formation: Implications for innate immunity. Blood 2011, 117, 953–959. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kernien, J.F.; Johnson, C.J.; Nett, J.E. Conserved Inhibition of Neutrophil Extracellular Trap Release by Clinical Candida albicans Biofilms. J. Fungi 2017, 3, 49. https://doi.org/10.3390/jof3030049
Kernien JF, Johnson CJ, Nett JE. Conserved Inhibition of Neutrophil Extracellular Trap Release by Clinical Candida albicans Biofilms. Journal of Fungi. 2017; 3(3):49. https://doi.org/10.3390/jof3030049
Chicago/Turabian StyleKernien, John F., Chad J. Johnson, and Jeniel E. Nett. 2017. "Conserved Inhibition of Neutrophil Extracellular Trap Release by Clinical Candida albicans Biofilms" Journal of Fungi 3, no. 3: 49. https://doi.org/10.3390/jof3030049