Diversity of Cell Wall Related Proteins in Human Pathogenic Fungi
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. The Dataset
3.2. Cell Wall Components
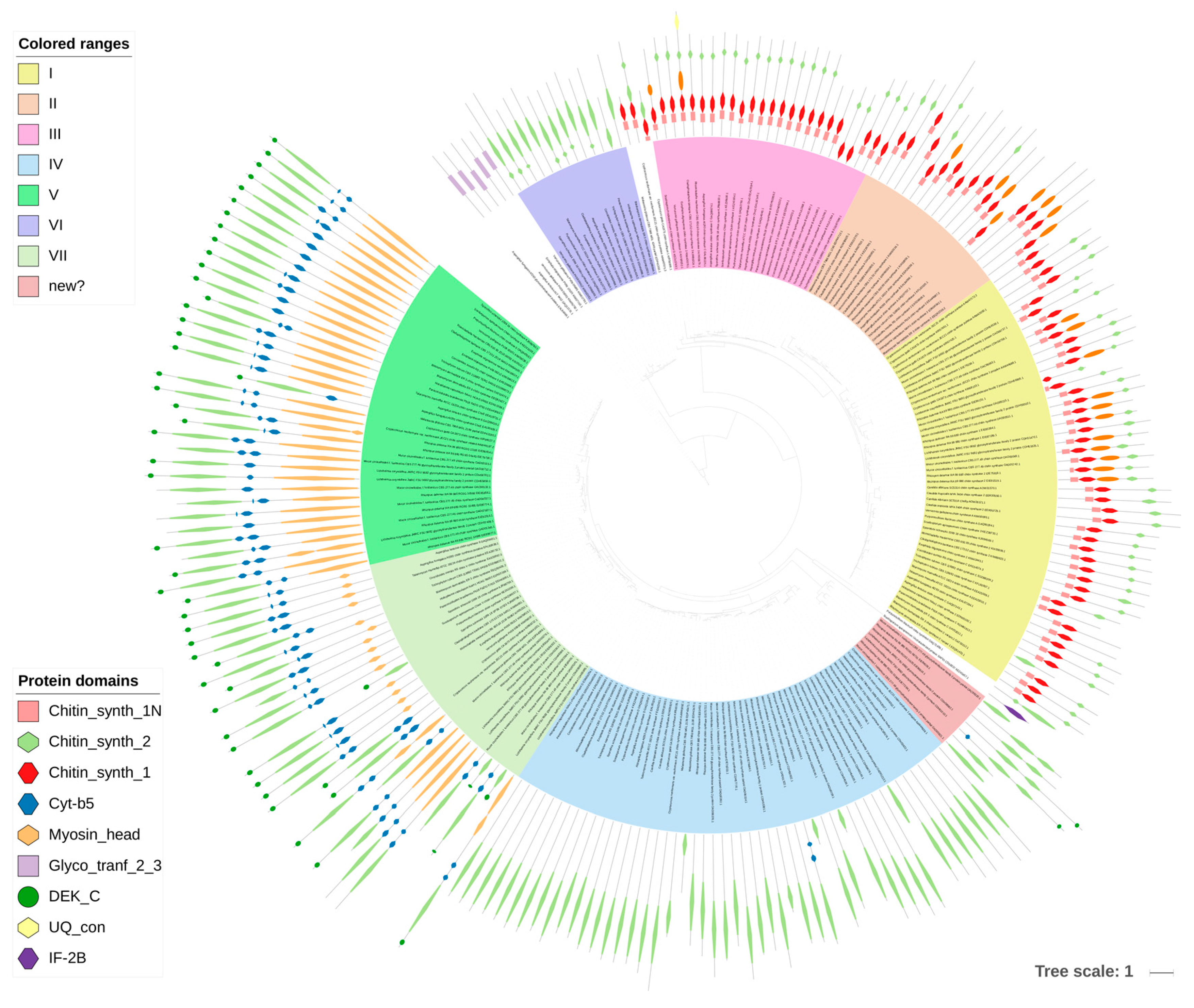
3.2.1. Chitin
3.2.2. Glucan
3.2.3. Glycosylation and Synthesis of Mannan
3.2.4. Other Cell Wall Components
3.3. Diverse Fungal Lineages
3.3.1. The Minimalists: Pneumocystis and Malassezia
3.3.2. The Complex Eurotiales
3.3.3. Chromoblastomycosis Causing Fungi
3.3.4. Animal-Related Onygenales
3.3.5. Opportunistic Sordariales
3.3.6. Commensal and Pathogenic—Candida
3.3.7. Cryptococcus and the Capsule
3.3.8. Mucoromycotina
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Erwig, L.P.; Gow, N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016, 14, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P.; Beauvais, A. Functional duality of the cell wall. Curr. Opin. Microbiol. 2014, 20, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Free, S.J. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013, 81, 33–82. [Google Scholar] [PubMed]
- Samar, D.; Kieler, J.B.; Klutts, J.S. Identification and deletion of Tft1, a predicted glycosyltransferase necessary for cell wall β-1,3;1,4-glucan synthesis in Aspergillus fumigatus. PLoS ONE 2015, 10, e0117336. [Google Scholar] [CrossRef] [PubMed]
- Hogan, L.H.; Klein, B.S.; Levitz, S.M. Virulence factors of medically important fungi. Clin. Microbiol. Rev. 1996, 9, 469–488. [Google Scholar] [PubMed]
- Latgé, J.-P.; Beauvais, A.; Chamilos, G. The cell wall of the human fungal pathogen Aspergillus fumigatus: Biosynthesis, organization, immune response, and virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Leonard, G.; Wideman, J.G. What defines the “kingdom” fungi? Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Armstrong-James, D.; Meintjes, G.; Brown, G.D. A neglected epidemic: Fungal infections in HIV/AIDS. Trends Microbiol. 2014, 22, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.S.D.; Dodds Ashley, E.S.; Lewis, R.; Lewis, J.S.; Martin, C.; Andes, D. Pharmacology of systemic antifungal agents. Clin. Infect. Dis. 2006, 43, S28–S39. [Google Scholar] [CrossRef]
- Cortez, K.J.; Roilides, E.; Quiroz-Telles, F.; Meletiadis, J.; Antachopoulos, C.; Knudsen, T.; Buchanan, W.; Milanovich, J.; Sutton, D.A.; Fothergill, A.; et al. Infections caused by Scedosporium spp. Clin. Microbiol. Rev. 2008, 21, 157–197. [Google Scholar] [CrossRef] [PubMed]
- Slavin, M.; van Hal, S.; Sorrell, T.C.; Lee, A.; Marriott, D.J.; Daveson, K.; Kennedy, K.; Hajkowicz, K.; Halliday, C.; Athan, E.; et al. Invasive infections due to filamentous fungi other than Aspergillus: Epidemiology and determinants of mortality. Clin. Microbiol. Infect. 2015, 21, 490.e1–490.e10. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Bromley, M.J. Infectious disease. How to bolster the antifungal pipeline. Science 2015, 347, 1414–1416. [Google Scholar] [CrossRef] [PubMed]
- Ncbi Resource Coordinators. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017, 45, D12–D17. [Google Scholar]
- Terrapon, N.; Lombard, V.; Drula, E.; Coutinho, P.M.; Henrissat, B. The CAZy database/the carbohydrate-active enzyme (CAZy) database: Principles and usage guidelines. In A Practical Guide to Using Glycomics Databases; Springer: Tokayo, Japan, 2016; pp. 117–131. [Google Scholar]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhang, H.; Wu, P.; Entwistle, S.; Li, X.; Yohe, T.; Yi, H.; Yang, Z.; Yin, Y. dbCAN-seq: A database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: Hmmer3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Oliveira da Silva, J.; da Silva, J.O.; Orellana, E.T.V.; Delgado, M.X.T. Development of a parallel version of phyml 3.0 using shared memory. IEEE Lat. Am. Trans. 2017, 15, 959–967. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Hoegger, P.J.; Kilaru, S.; James, T.Y.; Thacker, J.R.; Kues, U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006, 273, 2308–2326. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, M.; Kanehisa, M. Using the KEGG database resource. In Current Protocols in Bioinformatics; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A.R. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartnicki-Garcia, S. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu. Rev. Microbiol. 1968, 22, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Ibrahim, A.S.; Skory, C.; Grabherr, M.G.; Burger, G.; Butler, M.; Elias, M.; Idnurm, A.; Lang, B.F.; Sone, T.; et al. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009, 5, e1000549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mélida, H.; Sain, D.; Stajich, J.E.; Bulone, V. Deciphering the uniqueness of mucoromycotina cell walls by combining biochemical and phylogenomic approaches. Environ. Microbiol. 2015, 17, 1649–1662. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Berbee, M.L. No jacket required—New fungal lineage defies dress code. Bioessays 2011, 34, 94–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebé-Pedrós, A.; Grau-Bové, X.; Richards, T.A.; Ruiz-Trillo, I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 2014, 6, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Schmoll, M.; Dattenböck, C.; Carreras-Villaseñor, N.; Mendoza-Mendoza, A.; Tisch, D.; Alemán, M.I.; Baker, S.E.; Brown, C.; Cervantes-Badillo, M.G.; Cetz-Chel, J.; et al. The genomes of three uneven siblings: Footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 2016, 80, 205–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.M.; Gow, N.A.R.; Brown, G.D. Pattern recognition: Recent insights from dectin-1. Curr. Opin. Immunol. 2009, 21, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Rappleye, C.A.; Eissenberg, L.G.; Goldman, W.E. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Stalhberger, T.; Simenel, C.; Clavaud, C.; Eijsink, V.G.H.; Jourdain, R.; Delepierre, M.; Latgé, J.-P.; Breton, L.; Fontaine, T. Chemical organization of the cell wall polysaccharide core of Malassezia restricta. J. Biol. Chem. 2014, 289, 12647–12656. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Alspaugh, J.A. The Cryptococcus neoformans capsule: A sword and a shield. Clin. Microbiol. Rev. 2012, 25, 387–408. [Google Scholar] [CrossRef] [PubMed]
- Steczkiewicz, K.; Knizewski, L.; Rychlewski, L.; Ginalski, K. Tos1 is circularly permuted 1,3-β-glucanase. Cell Cycle 2010, 9, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Costachel, C.; Coddeville, B.; Latgé, J.-P.; Fontaine, T. Glycosylphosphatidylinositol-anchored fungal polysaccharide in Aspergillus fumigatus. J. Biol. Chem. 2005, 280, 39835–39842. [Google Scholar] [CrossRef] [PubMed]
- McMurrough, I.; Rose, A.H. Effect of growth rate and substrate limitation on the composition and structure of the cell wall of Saccharomyces cerevisiae. Biochem. J. 1967, 105, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Ovalle, R. Cell wall architecture in yeast: New structure and new challenges. J. Bacteriol. 1998, 180, 3735–3740. [Google Scholar] [PubMed]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Leal, J.A.; Jiménez-Barbero, J.; Gómez-Miranda, B.; Prieto, A.; Domenech, J.; Bernabé, M. Structural investigation of a cell-wall galactomannan from Neurospora crassa and N. sitophila. Carbohydr. Res. 1996, 283, 215–222. [Google Scholar] [CrossRef]
- Cantu, D.; Greve, L.C.; Labavitch, J.M.; Powell, A.L.T. Characterization of the cell wall of the ubiquitous plant pathogen botrytis cinerea. Mycol. Res. 2009, 113, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Morelle, W.; Bernard, M.; Debeaupuis, J.P.; Buitrago, M.; Tabouret, M.; Latgé, J.P. Galactomannoproteins of Aspergillus fumigatus. Eukaryot. Cell 2005, 4, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Aebi, M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta (BBA)—Gen. Subj. 1999, 1426, 239–257. [Google Scholar] [CrossRef]
- Lennarz, W.J. Studies on oligosaccharyl transferase in yeast. Acta Biochim. Pol. 2007, 54, 673–677. [Google Scholar] [PubMed]
- Munro, S. What can yeast tell us about N-linked glycosylation in the golgi apparatus? FEBS Lett. 2001, 498, 223–227. [Google Scholar] [CrossRef]
- Ruiz-Herrera, J.; Victoria Elorza, M.; Valentín, E.; Sentandreu, R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006, 6, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Gemmill, T.R.; Trimble, R.B. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1999, 1426, 227–237. [Google Scholar] [CrossRef]
- Lommel, M.; Strahl, S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology 2009, 19, 816–828. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Scheinost, A. Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 1999, 274, 9068–9075. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; White, A.M.; Sheraton, J.; di Paolo, T.; Treadwell, J.; Southard, S.B.; Horenstein, C.I.; Chen-Weiner, J.; Ram, A.F.; Kapteyn, J.C.; et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 1997, 147, 435–450. [Google Scholar] [PubMed]
- Lussier, M.; Sdicu, A.M.; Bussey, H. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1999, 1426, 323–334. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.F.; Mora-Montes, H.M.; Hernández-Cervantes, A.; Luna-Arias, J.P.; Gow, N.A.R.; Flores-Carreón, A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 2012, 419, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Bates, S.; Buurman, E.T.; Hughes, H.B.; Maccallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.J.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans are partially redundant α-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005, 280, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Díaz-Jiménez, D.F.; Jan Kullberg, B.; Brown, A.J.P.; Odds, F.C.; et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 2010, 285, 12087–12095. [Google Scholar] [CrossRef] [PubMed]
- Herscovics, A.; Orlean, P. Glycoprotein biosynthesis in yeast. FASEB J. 1993, 7, 540–550. [Google Scholar] [PubMed]
- Fontaine, T.; Magnin, T.; Melhert, A.; Lamont, D.; Latge, J.-P.; Ferguson, M.A.J. Structures of the glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology 2003, 13, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Lipke, P.N.; Kurjan, J. Sexual agglutination in budding yeasts: Structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 1992, 56, 180–194. [Google Scholar] [PubMed]
- Shahinian, S.; Dijkgraaf, G.J.; Sdicu, A.M.; Thomas, D.Y.; Jakob, C.A.; Aebi, M.; Bussey, H. Involvement of protein N-glycosyl chain glucosylation and processing in the biosynthesis of cell wall β-1,6-glucan of Saccharomyces cerevisiae. Genetics 1998, 149, 843–856. [Google Scholar] [PubMed]
- De Groot, P.W.J.; Ram, A.F.; Klis, F.M. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 2005, 42, 657–675. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Boorsma, A.; De Groot, P.W.J. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Lipke, P.N. On the evolution of fungal and yeast cell walls. Yeast 2010, 27, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Levery, S.B.; Momany, M.; Lindsey, R.; Toledo, M.S.; Shayman, J.A.; Fuller, M.; Brooks, K.; Doong, R.L.; Straus, A.H.; Takahashi, H.K. Disruption of the glucosylceramide biosynthetic pathway in Aspergillus nidulans and Aspergillus fumigatus by inhibitors of UDP-GIc:ceramide glucosyltransferase strongly affects spore germination, cell cycle, and hyphal growth. FEBS Lett. 2002, 525, 59–64. [Google Scholar] [CrossRef]
- Del Poeta, M.; Nimrichter, L.; Rodrigues, M.L.; Luberto, C. Synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 2014, 10, e1003832. [Google Scholar] [CrossRef] [PubMed]
- Nimrichter, L.; Rodrigues, M.L. Fungal glucosylceramides: From structural components to biologically active targets of new antimicrobials. Front. Microbiol. 2011, 2, 212. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Travassos, L.R.; Miranda, K.R.; Franzen, A.J.; Rozental, S.; de Souza, W.; Alviano, C.S.; Barreto-Bergter, E. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect. Immun. 2000, 68, 7049–7060. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.-Y.; Zhu, H.-M.; Wu, J.-H.; Wen, H.; Liu, C.-J. Increased permeability of blood-brain barrier is mediated by serine protease during Cryptococcus meningitis. J. Int. Med. Res. 2014, 42, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Komachi, Y.; Hatakeyama, S.; Motomatsu, H.; Futagami, T.; Kizjakina, K.; Sobrado, P.; Ekino, K.; Takegawa, K.; Goto, M.; Nomura, Y.; et al. Gfsa encodes a novel galactofuranosyltransferase involved in biosynthesis of galactofuranose antigen of O-glycan in Aspergillus nidulans and Aspergillus fumigatus. Mol. Microbiol. 2013, 90, 1054–1073. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Delangle, A.; Simenel, C.; Coddeville, B.; van Vliet, S.J.; van Kooyk, Y.; Bozza, S.; Moretti, S.; Schwarz, F.; Trichot, C.; et al. Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 2011, 7, e1002372. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Chan, C.M.; Lee, C.; Wong, S.S.; Yuen, K.Y. Mp1 encodes an abundant and highly antigenic cell wall mannoprotein in the pathogenic fungus Penicillium marneffei. Infect. Immun. 1998, 66, 966–973. [Google Scholar] [PubMed]
- Delattre, C.; Fenoradosoa, T.A.; Michaud, P. Galactans: An overview of their most important sourcing and applications as natural polysaccharides. Braz. Arch. Biol. Technol. 2011, 54, 1075–1092. [Google Scholar] [CrossRef]
- Lopes, L.C.L.; da Silva, M.I.D.; Bittencourt, V.C.B.; Figueiredo, R.T.; Rollin-Pinheiro, R.; Sassaki, G.L.; Bozza, M.T.; Gorin, P.A.J.; Barreto-Bergter, E. Glycoconjugates and polysaccharides from the Scedosporium/Pseudallescheria boydii complex: Structural characterisation, involvement in cell differentiation, cell recognition and virulence. Mycoses 2011, 54, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Salas, O.; Bowler, K.; Oren-Young, L.; Bar-Peled, M.; Sharon, A. Genetic alteration of udp-rhamnose metabolism inbotrytis cinerealeads to the accumulation of UDP-KDG that adversely affects development and pathogenicity. Mol. Plant Pathol. 2016, 18, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, P.; Boshoven, J.C.; Salas, O.; Bowler, K.; Islam, M.T.; Saber, M.K.; van den Berg, G.C.M.; Bar-Peled, M.; Thomma, B.P.H.J. Rhamnose synthase activity is required for pathogenicity of the vascular wilt fungus verticillium dahliae. Mol. Plant Pathol. 2017, 18, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Bento, I.; Martins, L.O.; Gato Lopes, G.; Arménia Carrondo, M.; Lindley, P.F. Dioxygen reduction by multi-copper oxidases; a structural perspective. Dalton Trans. 2005, 7, 3507–3513. [Google Scholar] [CrossRef] [PubMed]
- Frohner, I.E.; Bourgeois, C.; Yatsyk, K.; Majer, O.; Kuchler, K. Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol. Microbiol. 2009, 71, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Saunders, C.W.; Hu, P.; Grant, R.A.; Boekhout, T.; Kuramae, E.E.; Kronstad, J.W.; Deangelis, Y.M.; Reeder, N.L.; Johnstone, K.R.; et al. Dandruff-associated malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 2007, 104, 18730–18735. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, Z.; Huang, D.W.; Kutty, G.; Ishihara, M.; Wang, H.; Abouelleil, A.; Bishop, L.; Davey, E.; Deng, R.; et al. Genome analysis of three pneumocystis species reveals adaptation mechanisms to life exclusively in mammalian hosts. Nat. Commun. 2016, 7, 10740. [Google Scholar] [CrossRef] [PubMed]
- Banks, I.R.; Specht, C.A.; Donlin, M.J.; Gerik, K.J.; Levitz, S.M.; Lodge, J.K. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 2005, 4, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Gioti, A.; Nystedt, B.; Li, W.; Xu, J.; Andersson, A.; Averette, A.F.; Münch, K.; Wang, X.; Kappauf, C.; Kingsbury, J.M.; et al. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio 2013, 4, e00572-12. [Google Scholar] [CrossRef] [PubMed]
- Van de Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latgé, J.-P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Pasricha, S.; Payne, M.; Canovas, D.; Pase, L.; Ngaosuwankul, N.; Beard, S.; Oshlack, A.; Smyth, G.K.; Chaiyaroj, S.C.; Boyce, K.J.; et al. Cell-type-specific transcriptional profiles of the dimorphic pathogen Penicillium marneffei reflect distinct reproductive, morphological, and environmental demands. G3 2013, 3, 1997–2014. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, S.; Maeda, H.; Takahashi, T.; Yamagata, Y.; Hasegawa, F.; Gomi, K.; Nakajima, T.; Abe, K. Novel hydrophobic surface binding protein, HsbA, produced by Aspergillus oryzae. Appl. Environ. Microbiol. 2006, 72, 2407–2413. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Herrera-Estrella, A.; Seidl-Seiboth, V.; Martinez, D.A.; Druzhinina, I.S.; Thon, M.; Zeilinger, S.; Casas-Flores, S.; Horwitz, B.A.; Mukherjee, P.K.; et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of trichoderma. Genome Biol. 2011, 12, R40. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.M.; Moreno, L.F.; Stielow, B.J.; Muszewska, A.; Hainaut, M.; Gonzaga, L.; Abouelleil, A.; Patané, J.S.L.; Priest, M.; Souza, R.; et al. Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota). Stud. Mycol. 2017, 86, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, T.; Fujiyama, K.; Takegawa, K. Identification of novel α1,3-galactosyltransferase and elimination of α-galactose-containing glycans by disruption of multiple α-galactosyltransferase genes in Schizosaccharomyces pombe. J. Biol. Chem. 2012, 287, 38866–38875. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.F.; McEwen, J.G.; Clay, O.K.; Cuomo, C.A. Genome analysis reveals evolutionary mechanisms of adaptation in systemic dimorphic fungi. 2017. [Google Scholar] [CrossRef]
- Kanetsuna, F.; Carbonell, L.M. Cell wall composition of the yeastlike and mycelial forms of Blastomyces dermatitidis. J. Bacteriol. 1971, 106, 946–948. [Google Scholar] [PubMed]
- Kanetsuna, F.; Carbonell, L.M.; Moreno, R.E.; Rodriguez, J. Cell wall composition of the yeast and mycelial forms of Paracoccidioides brasiliensis. J. Bacteriol. 1969, 97, 1036–1041. [Google Scholar] [PubMed]
- Davis, T.E., Jr.; Domer, J.E.; Li, Y.T. Cell wall studies of Histoplasma capsulatum and Blastomyces dermatitidis using autologous and heterologous enzymes. Infect. Immun. 1977, 15, 978–987. [Google Scholar] [PubMed]
- Wheat, R.W.; Tritschler, C.; Conant, N.F.; Lowe, E.P. Comparison of Coccidioides immitis arthrospore, mycelium, and spherule cell walls, and influence of growth medium on mycelial cell wall composition. Infect. Immun. 1977, 17, 91–97. [Google Scholar] [PubMed]
- Goldman, C.; Akiyama, M.J.; Torres, J.; Louie, E.; Meehan, S.A. Scedosporium apiospermum infections and the role of combination antifungal therapy and GM-CSF: A case report and review of the literature. Med. Mycol. Case Rep. 2016, 11, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.M.; de Almeida, L.G.P.; Kubitschek-Barreira, P.; Alves, F.L.; Kioshima, E.S.; Abadio, A.K.R.; Fernandes, L.; Derengowski, L.S.; Ferreira, K.S.; Souza, R.C.; et al. Comparative genomics of the major fungal agents of human and animal sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis. BMC Genom. 2014, 15, 943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaffin, W.L. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008, 72, 495–544. [Google Scholar] [CrossRef] [PubMed]
- Cherniak, R.; Sundstrom, J.B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 1994, 62, 1507–1512. [Google Scholar] [PubMed]
- Reese, A.J.; Yoneda, A.; Breger, J.A.; Beauvais, A.; Liu, H.; Griffith, C.L.; Bose, I.; Kim, M.-J.; Skau, C.; Yang, S.; et al. Loss of cell wall α(1-3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 2007, 63, 1385–1398. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muszewska, A.; Piłsyk, S.; Perlińska-Lenart, U.; Kruszewska, J.S. Diversity of Cell Wall Related Proteins in Human Pathogenic Fungi. J. Fungi 2018, 4, 6. https://doi.org/10.3390/jof4010006
Muszewska A, Piłsyk S, Perlińska-Lenart U, Kruszewska JS. Diversity of Cell Wall Related Proteins in Human Pathogenic Fungi. Journal of Fungi. 2018; 4(1):6. https://doi.org/10.3390/jof4010006
Chicago/Turabian StyleMuszewska, Anna, Sebastian Piłsyk, Urszula Perlińska-Lenart, and Joanna S. Kruszewska. 2018. "Diversity of Cell Wall Related Proteins in Human Pathogenic Fungi" Journal of Fungi 4, no. 1: 6. https://doi.org/10.3390/jof4010006




