Bioengineering Microgels and Hydrogel Microparticles for Sensing Biomolecular Targets
Abstract
:1. Introduction
2. Hydrogels: Macrogels, Microparticles and Microgels
2.1. Synthesis and Modification of Microgels and Hydrogel Microparticles
2.1.1. Batch Synthesis of Microgels
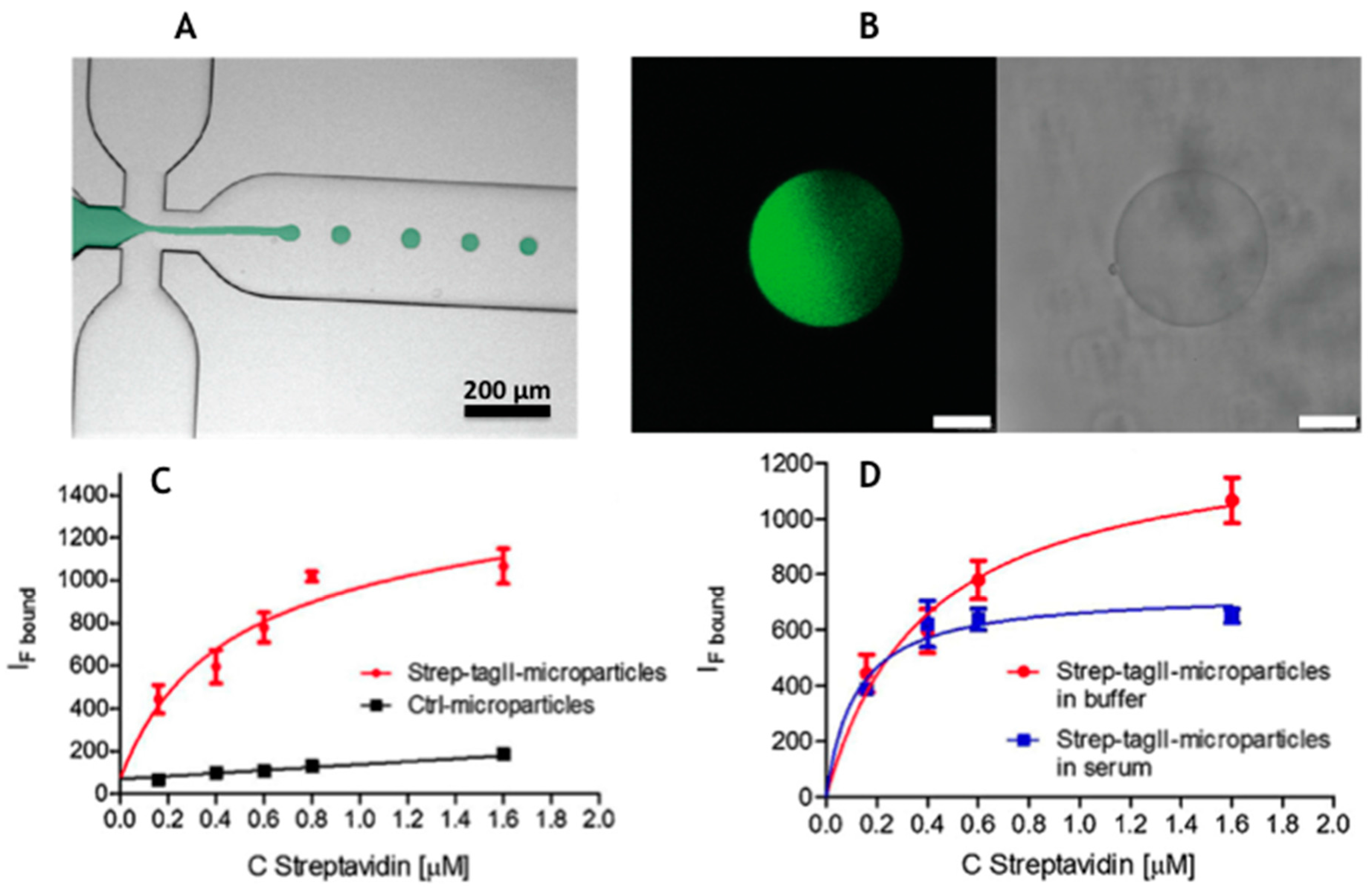
2.1.2. High Throughput Production of Hydrogel Microparticles by Microfluidics
2.2. Chemical Modifications
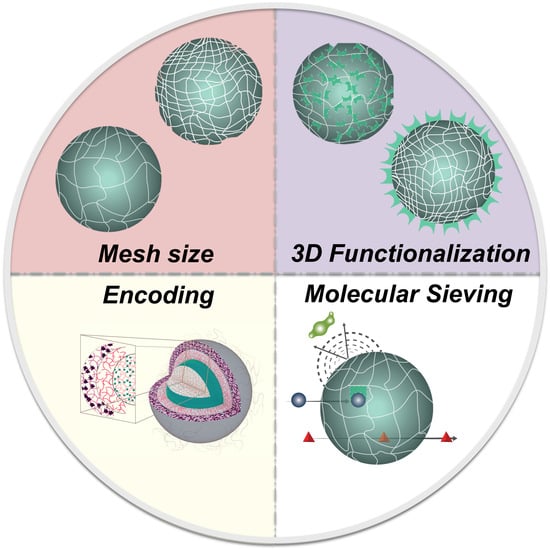
2.3. Hydrogel Characteristics for Biosensing
2.3.1. Swelling Behavior and Porosity
2.3.2. Probe Density
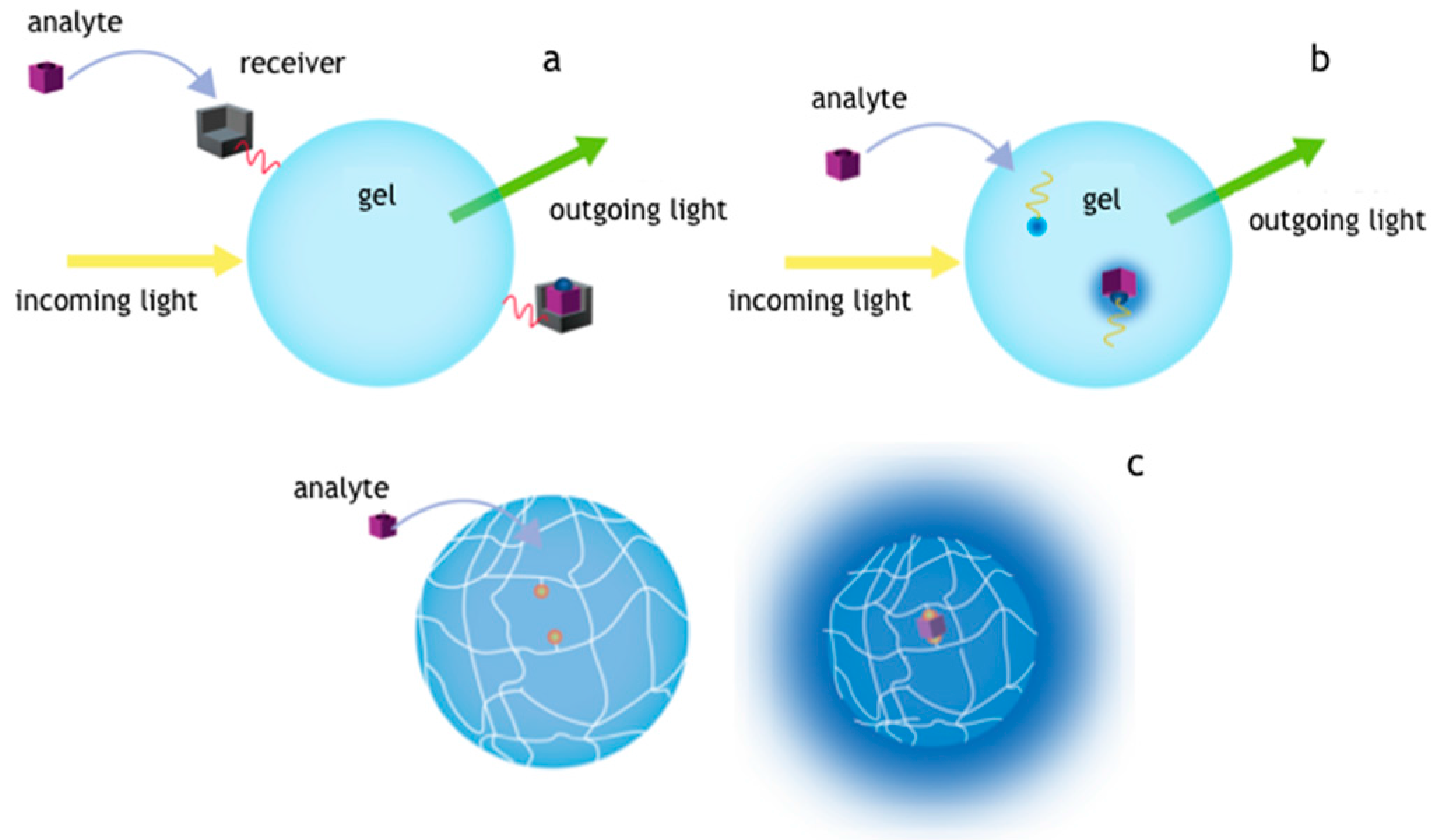
2.3.3. Target Diffusion in Hydrogel
2.3.4. Mass Transport Equations
3. Microgels and Hydrogel Microparticles in Multiplex Assays
3.1. Liquid ELISAs : Antibody Based Detction
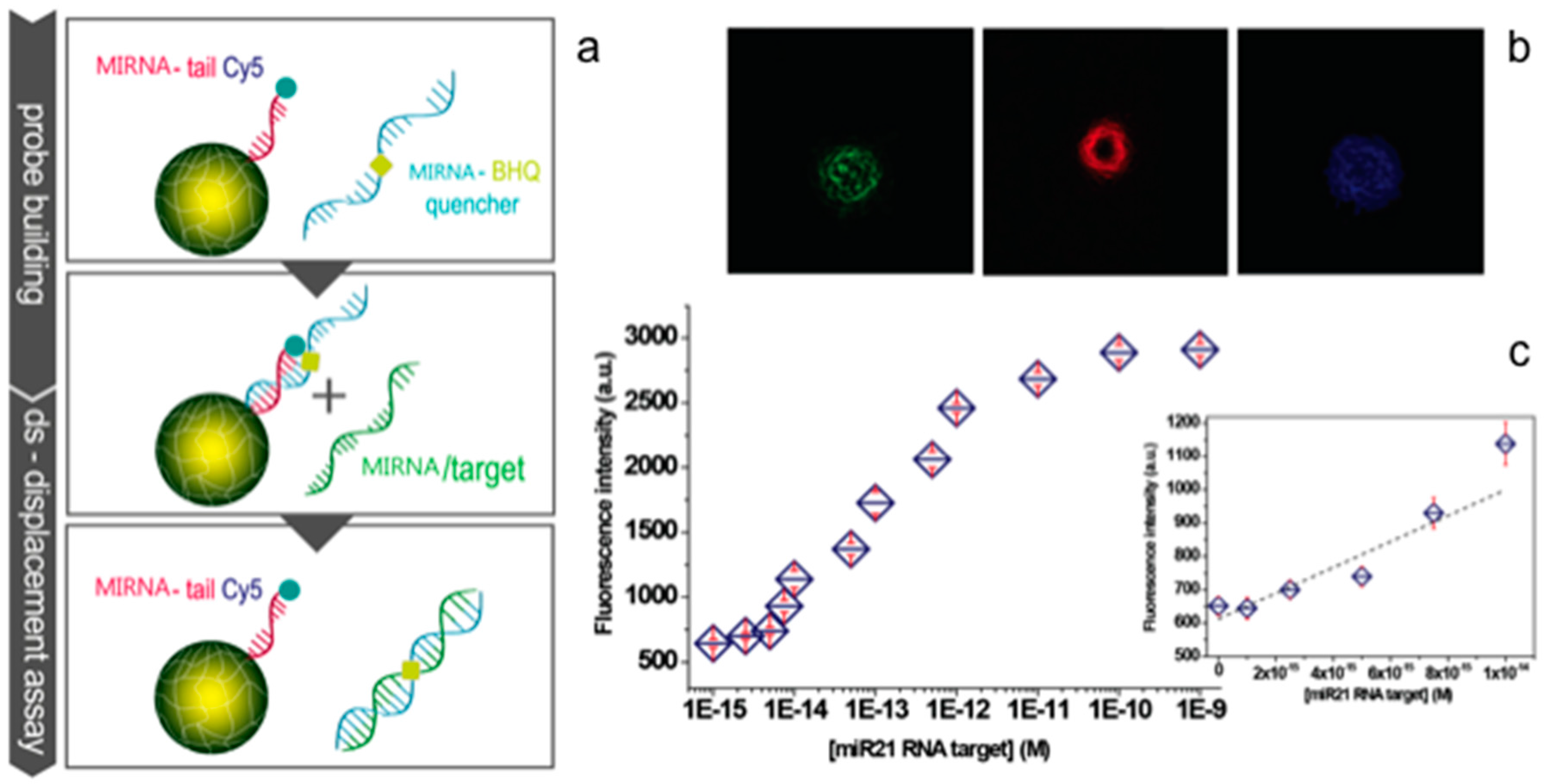
3.2. Oligonucleotides: MicroRNA and ssDNA
4. Conclusions and Outlooks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wilson, A.N.; Guiseppi-Elie, A. Bioresponsive hydrogels. Adv. Healthc. Mater. 2013, 2, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Kang, D.; Xue, Y.; Lee, S.; Pielak, R.M.; Kim, J.; Hwang, T.; Min, S.; Banks, A.; Bastien, P.; et al. A soft, wearable microfluidic device for the capture, storage, and colorimetric sensing of sweat. Sci. Transl. Med. 2016, 8, 366ra165. [Google Scholar] [CrossRef] [PubMed]
- Pregibon, D.C.; Toner, M.; Doyle, P.S. Multifunctional encoded particles for high-throughput biomolecule analysis. Science 2007, 315, 1393–1396. [Google Scholar] [CrossRef] [PubMed]
- Birtwell, S.; Morgan, H. Microparticle encoding technologies for high-throughput multiplexed suspension assays. Integr. Biol. (Camb.) 2009, 1, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Bawazer, L.A.; McNally, C.S.; Empson, C.J.; Marchant, W.J.; Comyn, T.P.; Niu, X.; Cho, S.; McPherson, M.J.; Binks, B.P.; deMello, A.; et al. Combinatorial microfluidic droplet engineering for biomimetic material synthesis. Sci. Adv. 2016, 2, e1600567. [Google Scholar] [CrossRef] [PubMed]
- Manikas, A.C.; Aliberti, A.; Causa, F.; Battista, E.; Netti, P.A. Thermoresponsive pnipaam hydrogel scaffolds with encapsulated aunps show high analyte-trapping ability and tailored plasmonic properties for high sensing efficiency. J. Mater. Chem. B 2015, 3, 53–58. [Google Scholar] [CrossRef]
- Appleyard, D.C.; Chapin, S.C.; Doyle, P.S. Multiplexed protein quantification with barcoded hydrogel microparticles. Anal. Chem. 2011, 83, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Cusano, A.M.; Causa, F.; Moglie, R.D.; Falco, N.; Scognamiglio, P.L.; Aliberti, A.; Vecchione, R.; Battista, E.; Marasco, D.; Savarese, M.; et al. Integration of binding peptide selection and multifunctional particles as tool-box for capture of soluble proteins in serum. J. R. Soc. Interface 2014, 11, 20140718. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Cossins, A.R.; Spiller, D.G. Encoded microcarriers for high-throughput multiplexed detection. Angew. Chem. Int. Ed. Engl. 2006, 45, 6104–6117. [Google Scholar] [CrossRef] [PubMed]
- Squires, T.M.; Messinger, R.J.; Manalis, S.R. Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 2008, 26, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Nanomaterial-based amplified transduction of biomolecular interactions. Small 2005, 1, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Qin, Y.; He, Y.; Huang, Q.; Fan, C.; Chen, H.Y. Functional nanoprobes for ultrasensitive detection of biomolecules. Chem. Soc. Rev. 2010, 39, 4234–4243. [Google Scholar] [CrossRef] [PubMed]
- Wilner, O.I.; Willner, B.; Willner, I. DNA nanotechnology. Adv. Exp. Med. Biol. 2012, 733, 97–114. [Google Scholar] [PubMed]
- Li, Y.; Luo, D. Multiplexed molecular detection using encoded microparticles and nanoparticles. Expert Rev. Mol. Diagn. 2006, 6, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.H.; Ilag, L.L. Biochips beyond DNA: Technologies and applications. Biotechnol. Annu. Rev. 2003, 9, 1–149. [Google Scholar] [PubMed]
- Wu, W.; Zhou, S. Hybrid micro-/nanogels for optical sensing and intracellular imaging. Nano Rev. 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Waggoner, P.S.; Craighead, H.G. Micro- and nanomechanical sensors for environmental, chemical, and biological detection. Lab Chip 2007, 7, 1238–1255. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B.R.; Govindhan, M.; Chen, A. Carbon nanomaterials based electrochemical sensors/biosensors for the sensitive detection of pharmaceutical and biological compounds. Sensors (Basel) 2015, 15, 22490–22508. [Google Scholar] [CrossRef] [PubMed]
- Peppas, N.A.; Bures, P.; Leobandung, W.; Ichikawa, H. Hydrogels in pharmaceutical formulations. Eur. J. Pharm. Biopharm. 2000, 50, 27–46. [Google Scholar] [CrossRef]
- Nayak, S.; Lyon, L.A. Ligand-functionalized core/shell microgels with permselective shells. Angew. Chem. Int. Ed. Engl. 2004, 43, 6706–6709. [Google Scholar] [CrossRef] [PubMed]
- Hoare, T.; Pelton, R. Functionalized microgel swelling: Comparing theory and experiment. J. Phys. Chem. B 2007, 111, 11895–11906. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.H.; Lyon, L.A. Tunable encapsulation of proteins within charged microgels. Macromolecules 2011, 44, 8154–8160. [Google Scholar] [CrossRef] [PubMed]
- Hoare, T.; Pelton, R. Dimensionless plot analysis: A new way to analyze functionalized microgels. J. Colloid Interface Sci. 2006, 303, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Tong, Z.; Lyon, L.A. Multicompartment core/shell microgels. J. Am. Chem. Soc. 2010, 132, 11470–11472. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.H.; Herman, E.S.; Lyon, L.A. Network deconstruction reveals network structure in responsive microgels. J. Phys. Chem. B 2011, 115, 3761–3764. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, G.C.; Srinivas, R.L.; Hill, W.A.; Doyle, P.S. Hydrogel microparticles for biosensing. Eur. Polym. J. 2015, 72, 386–412. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Martin, B.; Laporte, Y.; South, A.B.; Lyon, L.A.; Fernandez-Nieves, A. Bulk modulus of poly(N-isopropylacrylamide) microgels through the swelling transition. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2011, 84, 011406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.M.; Xu, W.; Serpe, M.J. Optical devices constructed from multiresponsive microgels. Angew. Chem. Int. Ed. Engl. 2014, 53, 4827–4831. [Google Scholar] [CrossRef] [PubMed]
- Clara-Rahola, J.; Fernandez-Nieves, A.; Sierra-Martin, B.; South, A.B.; Lyon, L.A.; Kohlbrecher, J.; Fernandez Barbero, A. Structural properties of thermoresponsive poly(N-isopropylacrylamide)-poly(ethyleneglycol) microgels. J. Chem. Phys. 2012, 136, 214903. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Gan, D.; Serpe, M.J.; Lyon, L.A. Hollow thermoresponsive microgels. Small 2005, 1, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Tong, Z.; Lyon, L.A. Synthesis and physicochemical properties of cationic microgels based on poly(N-isopropylmethacrylamide). Colloid Polym. Sci. 2010, 289, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Sun, B.; Li, X.; Yu, Y.; Tian, Y.; Xu, X.; Jin, Z. Synthesis of ph- and ionic strength-responsive microgels and their interactions with lysozyme. Int. J. Biol. Macromol. 2015, 79, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, D.; Maduar, S.R.; Santer, M.; Lomadze, N.; Vinogradova, O.I.; Santer, S. Manipulation of small particles at solid liquid interface: Light driven diffusioosmosis. Sci. Rep. 2016, 6, 36443. [Google Scholar] [CrossRef] [PubMed]
- Crassous, J.J.; Demirors, A.F. Multiscale directed self-assembly of composite microgels in complex electric fields. Soft Matter 2016, 13, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Barbero, A.; Suarez, I.J.; Sierra-Martin, B.; Fernandez-Nieves, A.; de Las Nieves, F.J.; Marquez, M.; Rubio-Retama, J.; Lopez-Cabarcos, E. Gels and microgels for nanotechnological applications. Adv. Colloid Interface Sci. 2009, 147–148, 88–108. [Google Scholar] [CrossRef] [PubMed]
- Battista, E.; Mazzarotta, A.; Causa, F.; Cusano, A.M.; Netti, P.A. Core—shell microgels with controlled structural properties. Polym. Int. 2016, 65, 747–755. [Google Scholar] [CrossRef]
- Bachman, H.; Brown, A.C.; Clarke, K.C.; Dhada, K.S.; Douglas, A.; Hansen, C.E.; Herman, E.; Hyatt, J.S.; Kodlekere, P.; Meng, Z.; et al. Ultrasoft, highly deformable microgels. Soft Matter 2015, 11, 2018–2028. [Google Scholar] [CrossRef] [PubMed]
- Causa, F.; Aliberti, A.; Cusano, A.M.; Battista, E.; Netti, P.A. Supramolecular spectrally encoded microgels with double strand probes for absolute and direct mirna fluorescence detection at high sensitivity. J. Am. Chem. Soc. 2015, 137, 1758–1761. [Google Scholar] [CrossRef] [PubMed]
- Celetti, G.; Natale, C.D.; Causa, F.; Battista, E.; Netti, P.A. Functionalized poly(ethylene glycol) diacrylate microgels by microfluidics: In situ peptide encapsulation for in serum selective protein detection. Colloids Surf. B 2016, 145, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.L.; Lin, Y.; Yang, C.; Manocchi, A.K.; Yuet, K.P.; Doyle, P.S.; Yi, H. Microfluidic fabrication of hydrogel microparticles containing functionalized viral nanotemplates. Langmuir 2010, 26, 13436–13441. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Bisso, P.W.; Srinivas, R.L.; Kim, J.J.; Swiston, A.J.; Doyle, P.S. Universal process-inert encoding architecture for polymer microparticles. Nat. Mater. 2014, 13, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Serra, C.A.; Chang, Z. Microfluidic-assisted synthesis of polymer particles. Chem. Eng. Technol. 2008, 31, 1099–1115. [Google Scholar] [CrossRef]
- Dendukuri, D.; Doyle, P.S. The synthesis and assembly of polymeric microparticles using microfluidics. Adv. Mater. 2009, 21, 4071–4086. [Google Scholar] [CrossRef]
- Wang, J.T.; Wang, J.; Han, J.J. Fabrication of advanced particles and particle-based materials assisted by droplet-based microfluidics. Small 2011, 7, 1728–1754. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.L.; Choi, C.H.; Lin, Y.; Lee, C.S.; Yi, H. Fabrication of uniform DNA-conjugated hydrogel microparticles via replica molding for facile nucleic acid hybridization assays. Anal. Chem. 2010, 82, 5851–5858. [Google Scholar] [CrossRef] [PubMed]
- Heida, T.; Neubauer, J.W.; Seuss, M.; Hauck, N.; Thiele, J.; Fery, A. Mechanically defined microgels by droplet microfluidics. Macromol. Chem. Phys. 2017, 218, 1600418. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, L. Passive and active droplet generation with microfluidics: A review. Lab Chip 2017, 17, 34–75. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.-H.; Jung, J.-H.; Hwang, T.-S.; Lee, C.-S. In situ microfluidic synthesis of monodisperse peg microspheres. Macromol. Res. 2009, 17, 163–167. [Google Scholar] [CrossRef]
- Dendukuri, D.; Pregibon, D.C.; Collins, J.; Hatton, T.A.; Doyle, P.S. Continuous-flow lithography for high-throughput microparticle synthesis. Nat. Mater. 2006, 5, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Nie, Z.; Seo, M.; Lewis, P.; Kumacheva, E.; Stone, H.A.; Garstecki, P.; Weibel, D.B.; Gitlin, I.; Whitesides, G.M. Generation of monodisperse particles by using microfluidics: Control over size, shape, and composition. Angew. Chem. Int. Ed. Engl. 2005, 44, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Stenger, R.J.; Mosbach, E.H.; Cohen, B.I.; Ayyad, N.; Wilson, A.M. Structural alterations in the gallbladder epithelium of hamsters and prairie dogs fed lithogenic diets. Hepatology 1994, 20, A403. [Google Scholar]
- Nie, Z.; Xu, S.; Seo, M.; Lewis, P.C.; Kumacheva, E. Polymer particles with various shapes and morphologies produced in continuous microfluidic reactors. J. Am. Chem. Soc. 2005, 127, 8058–8063. [Google Scholar] [CrossRef] [PubMed]
- Appleyard, D.C.; Chapin, S.C.; Srinivas, R.L.; Doyle, P.S. Bar-coded hydrogel microparticles for protein detection: Synthesis, assay and scanning. Nat. Protoc. 2011, 6, 1761–1774. [Google Scholar] [CrossRef] [PubMed]
- Causa, F.; Aliberti, A.; Cusano, A.M.; Battista, E.; Netti, P.A. Microgels for multiplex and direct fluorescence detection. In Proceedings of the Optical Methods for Inspection, Characterization, and Imaging of Biomaterials II, Munich, Germany, 22–24 June 2015; p. 952919. [Google Scholar]
- Aliberti, A.; Cusano, A.M.; Battista, E.; Causa, F.; Netti, P.A. High sensitive and direct fluorescence detection of single viral DNA sequences by integration of double strand probes onto microgels particles. Analyst 2016, 141, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Kryscio, D.R.; Peppas, N.A. Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater. 2012, 8, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Anseth, K.S.; Bowman, C.N.; Brannon-Peppas, L. Mechanical properties of hydrogels and their experimental determination. Biomaterials 1996, 17, 1647–1657. [Google Scholar] [CrossRef]
- Brannon-Peppas, L.; Peppas, N.A. Equilibrium swelling behavior of pH-sensitive hydrogels. Chem. Eng. Sci. 1991, 46, 715–722. [Google Scholar] [CrossRef]
- Kofinas, P.; Athanassiou, V.; Merrill, E.W. Hydrogels prepared by electron irradiation of poly(ethylene oxide) in water solution: Unexpected dependence of cross-link density and protein diffusion coefficients on initial PEO molecular weight. Biomaterials 1996, 17, 1547–1550. [Google Scholar] [CrossRef]
- Marek, S.R.; Conn, C.A.; Peppas, N.A. Cationic nanogels based on diethylaminoethyl methacrylate. Polymer 2010, 51, 1237–1243. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, C.; Peppas, N.A.; Mijangos, C. Novel strategy for the determination of UCST-like microgels network structure: Effect on swelling behavior and rheology. Soft Matter 2012, 8, 337–346. [Google Scholar] [CrossRef]
- Thakur, A.; Wanchoo, R.; Singh, P. Structural parameters and swelling behavior of pH sensitive poly(acrylamide-co-acrylic acid) hydrogels. Chem. Biochem. Eng. Q. 2011, 25, 181–194. [Google Scholar]
- Zustiak, S.P.; Leach, J.B. Hydrolytically degradable poly(ethylene glycol) hydrogel scaffolds with tunable degradation and mechanical properties. Biomacromolecules 2010, 11, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Malana, M.A.; Firdous, G.; Javed, A.; Zafar, Z.I.; Khan, M.S. Swelling kinetics and rheological characterization of methyl acrylate-vinyl acetate-acrylic acid hydrogels. Asian J. Chem. 2014, 26, 1898. [Google Scholar]
- Zubtsov, D.A.; Savvateeva, E.N.; Rubina, A.Y.; Pan‘kov, S.V.; Konovalova, E.V.; Moiseeva, O.V.; Chechetkin, V.R.; Zasedatelev, A.S. Comparison of surface and hydrogel-based protein microchips. Anal. Biochem. 2007, 368, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Zubtsov, D.A.; Ivanov, S.M.; Rubina, A.Y.; Dementieva, E.I.; Chechetkin, V.R.; Zasedatelev, A.S. Effect of mixing on reaction-diffusion kinetics for protein hydrogel-based microchips. J. Biotechnol. 2006, 122, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Tennico, Y.H.; Hutanu, D.; Koesdjojo, M.T.; Bartel, C.M.; Remcho, V.T. On-chip aptamer-based sandwich assay for thrombin detection employing magnetic beads and quantum dots. Anal. Chem. 2010, 82, 5591–5597. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, R.L.; Johnson, S.D.; Doyle, P.S. Oil-isolated hydrogel microstructures for sensitive bioassays on-chip. Anal. Chem. 2013, 85, 12099–12107. [Google Scholar] [CrossRef] [PubMed]
- Ramanujan, S.; Pluen, A.; McKee, T.D.; Brown, E.B.; Boucher, Y.; Jain, R.K. Diffusion and convection in collagen gels: Implications for transport in the tumor interstitium. Biophys. J. 2002, 83, 1650–1660. [Google Scholar] [CrossRef]
- Feldman, D. The theory of polymer dynamics, by M. Doi and S. F. Edwards, the clarendon press, oxford university press, new york, 1986, 391 pp. Price: $78.50. J. Polym. Sci. C Polym. Lett. 1989, 27, 239–240. [Google Scholar] [CrossRef]
- Pregibon, D.C.; Doyle, P.S. Optimization of encoded hydrogel particles for nucleic acid quantification. Anal. Chem. 2009, 81, 4873–4881. [Google Scholar] [CrossRef] [PubMed]
- Ogston, A.G.; Preston, B.N.; Wells, J.D. On the transport of compact particles through solutions of chain-polymers. Proc. R. Soc. Lond. A Math. Phys. Sci. 1973, 333, 297–316. [Google Scholar] [CrossRef]
- Walton, S.P.; Stephanopoulos, G.N.; Yarmush, M.L.; Roth, C.M. Thermodynamic and kinetic characterization of antisense oligodeoxynucleotide binding to a structured mrna. Biophys. J. 2002, 82, 366–377. [Google Scholar] [CrossRef]
- Kastritis, P.L.; Bonvin, A.M. On the binding affinity of macromolecular interactions: Daring to ask why proteins interact. J. R. Soc. Interface 2013, 10, 20120835. [Google Scholar] [CrossRef] [PubMed]
- Spiga, O.; Bernini, A.; Scarselli, M.; Ciutti, A.; Bracci, L.; Lozzi, L.; Lelli, B.; di Maro, D.; Calamandrei, D.; Niccolai, N. Peptide-protein interactions studied by surface plasmon and nuclear magnetic resonances. FEBS Lett. 2002, 511, 33–35. [Google Scholar] [CrossRef]
- Zhao, Y.; Cheng, Y.; Shang, L.; Wang, J.; Xie, Z.; Gu, Z. Microfluidic synthesis of barcode particles for multiplex assays. Small 2015, 11, 151–174. [Google Scholar] [CrossRef] [PubMed]
- Oleinikov, V.A. Semiconductor fluorescent nanocrystals (quantum dots) in biological biochips. Bioorg. Khim. 2011, 37, 171–189. [Google Scholar] [PubMed]
- Han, M.; Gao, X.; Su, J.Z.; Nie, S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001, 19, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, X.; Li, Y.; Shi, Y.; Su, X. Multicolor quantum dot-encoded microspheres for the detection of biomolecules. Talanta 2007, 72, 1446–1452. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.X.; Chen, B.A.; Wang, H.; Xia, G.H.; Cheng, J.; Pei, X.P.; Wang, F.; Bao, W. Handy, rapid and multiplex detection of tumor markers based on encoded silica-hydrogel hybrid beads array chip. Biosens. Bioelectron. 2013, 48, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Yeom, S.Y.; Son, C.H.; Kim, B.S.; Tag, S.H.; Nam, E.; Shin, H.; Kim, S.H.; Gang, H.; Lee, H.J.; Choi, J.; et al. Multiplexed detection of epigenetic markers using quantum dot (QD)-encoded hydrogel microparticles. Anal. Chem. 2016, 88, 4259–4268. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mrna translation and stability by micrornas. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of micrornas in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Goretti, E.; Devaux, Y. Which future for circulating micrornas as biomarkers of acute myocardial infarction? Ann. Transl. Med. 2016, 4, 440. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Song, Y.; Yao, L.; Song, G.; Teng, C. Circulating micrornas: Promising biomarkers involved in several cancers and other diseases. DNA Cell Biol. 2017, 36, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, J.; Ochiya, T. Circulating micrornas and extracellular vesicles as potential cancer biomarkers: A systematic review. Int. J. Clin. Oncol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.N.; Nalpas, N.C.; McLoughlin, K.E.; Browne, J.A.; Gordon, S.V.; MacHugh, D.E.; Shaughnessy, R.G. Circulating micrornas as potential biomarkers of infectious disease. Front. Immunol. 2017, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C. Real-time liquid biopsy in cancer patients: Fact or fiction? Cancer Res. 2013, 73, 6384–6388. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C. Liquid biopsy: Potential and challenges. Mol. Oncol. 2016, 10, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, M.; Khatamianfar, S.; Hassanian, S.M.; Nedaeinia, R.; Shafiee, M.; Maftouh, M.; Ghayour-Mobarhan, M.; Sales, S.S.; Avan, A. Exosome-encapsulated micrornas as potential circulating biomarkers in colon cancer. Curr. Pharm. Des. 2017, 23, 1705–1709. [Google Scholar] [CrossRef]
- Hou, J.; Meng, F.; Chan, L.W.; Cho, W.C.; Wong, S.C. Circulating plasma micrornas as diagnostic markers for nsclc. Front. Genet. 2016, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lin, J.; Kong, D.; Huang, M.; Xu, C.; Kim, T.K.; Etheridge, A.; Luo, Y.; Ding, Y.; Wang, K. Current state of circulating micrornas as cancer biomarkers. Clin. Chem. 2015, 61, 1138–1155. [Google Scholar] [CrossRef] [PubMed]
- Erener, S.; Marwaha, A.; Tan, R.; Panagiotopoulos, C.; Kieffer, T.J. Profiling of circulating micrornas in children with recent onset of type 1 diabetes. JCI Insight 2017, 2, e89656. [Google Scholar] [CrossRef] [PubMed]
- Guay, C.; Regazzi, R. Circulating micrornas as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol 2013, 9, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Romaine, S.P.; Tomaszewski, M.; Condorelli, G.; Samani, N.J. Micrornas in cardiovascular disease: An introduction for clinicians. Heart 2015, 101, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Wang, D.Z. The emerging role of micrornas as a therapeutic target for cardiovascular disease. BioDrugs 2010, 24, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Maegdefessel, L. The emerging role of micrornas in cardiovascular disease. J. Intern. Med. 2014, 276, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Lee, E.J.; Jiang, J.; Sarkar, A.; Yang, L.; Elton, T.S.; Chen, C. Real-time pcr quantification of precursor and mature microrna. Methods 2008, 44, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011, 3, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Herrmann, M.G.; Gundry, C.N.; Elenitoba-Johnson, K.S. Real-time multiplex PCR assays. Methods 2001, 25, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Chapin, S.C.; Appleyard, D.C.; Pregibon, D.C.; Doyle, P.S. Rapid microrna profiling on encoded gel microparticles. Angew. Chem. Int. Ed. Engl. 2011, 50, 2289–2293. [Google Scholar] [CrossRef] [PubMed]
- Chapin, S.C.; Doyle, P.S. Ultrasensitive multiplexed microrna quantification on encoded gel microparticles using rolling circle amplification. Anal. Chem. 2011, 83, 7179–7185. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Jiang, L. Bio-inspired, smart, multiscale interfacial materials. Adv. Mater. 2008, 20, 2842–2858. [Google Scholar] [CrossRef]
- Dannhauser, D.; Causa, F.; Battista, E.; Cusano, A.M.; Rossi, D.; Netti, P.A. In-flow real-time detection of spectrally encoded microgels for mirna absolute quantification. Biomicrofluidics 2016, 10, 064114. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.J.; Ober, T.J.; Edd, J.F.; Desai, S.P.; Neal, D.; Bong, K.W.; Doyle, P.S.; McKinley, G.H.; Toner, M. Inertio-elastic focusing of bioparticles in microchannels at high throughput. Nat. Commun. 2014, 5, 4120. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Lee, S.S.; Hyun, K.; Lee, S.J.; Kim, J.M. DNA-based highly tunable particle focuser. Nat. Commun. 2013, 4, 2567. [Google Scholar] [CrossRef] [PubMed]
- Romeo, G.; D’Avino, G.; Greco, F.; Netti, P.A.; Maffettone, P.L. Viscoelastic flow-focusing in microchannels: Scaling properties of the particle radial distributions. Lab Chip 2013, 13, 2802–2807. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, G.; Romeo, G.; Villone, M.M.; Greco, F.; Netti, P.A.; Maffettone, P.L. Single line particle focusing induced by viscoelasticity of the suspending liquid: Theory, experiments and simulations to design a micropipe flow-focuser. Lab Chip 2012, 12, 1638–1645. [Google Scholar] [CrossRef] [PubMed]
- Del Giudice, F.; D'Avino, G.; Greco, F.; de Santo, I.; Netti, P.A.; Maffettone, P.L. Rheometry-on-a-chip: Measuring the relaxation time of a viscoelastic liquid through particle migration in microchannel flows. Lab Chip 2015, 15, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Dannhauser, D.; Romeo, G.; Causa, F.; de Santo, I.; Netti, P.A. Multiplex single particle analysis in microfluidics. Analyst 2014, 139, 5239–5246. [Google Scholar] [CrossRef] [PubMed]
- Dannhauser, D.; Rossi, D.; Causa, F.; Memmolo, P.; Finizio, A.; Wriedt, T.; Hellmers, J.; Eremin, Y.; Ferraro, P.; Netti, P.A. Optical signature of erythrocytes by light scattering in microfluidic flows. Lab Chip 2015, 15, 3278–3285. [Google Scholar] [CrossRef] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battista, E.; Causa, F.; Netti, P.A. Bioengineering Microgels and Hydrogel Microparticles for Sensing Biomolecular Targets. Gels 2017, 3, 20. https://doi.org/10.3390/gels3020020
Battista E, Causa F, Netti PA. Bioengineering Microgels and Hydrogel Microparticles for Sensing Biomolecular Targets. Gels. 2017; 3(2):20. https://doi.org/10.3390/gels3020020
Chicago/Turabian StyleBattista, Edmondo, Filippo Causa, and Paolo Antonio Netti. 2017. "Bioengineering Microgels and Hydrogel Microparticles for Sensing Biomolecular Targets" Gels 3, no. 2: 20. https://doi.org/10.3390/gels3020020