Regulation of the U3-, U8-, and U13snoRNA Expression by the DEAD Box Proteins Ddx5/Ddx17 with Consequences for Cell Proliferation and Survival
Abstract
:1. Introduction
2. Results
2.1. Ddx5 Regulates the Expression of U3-, U8-, and U13snoRNA
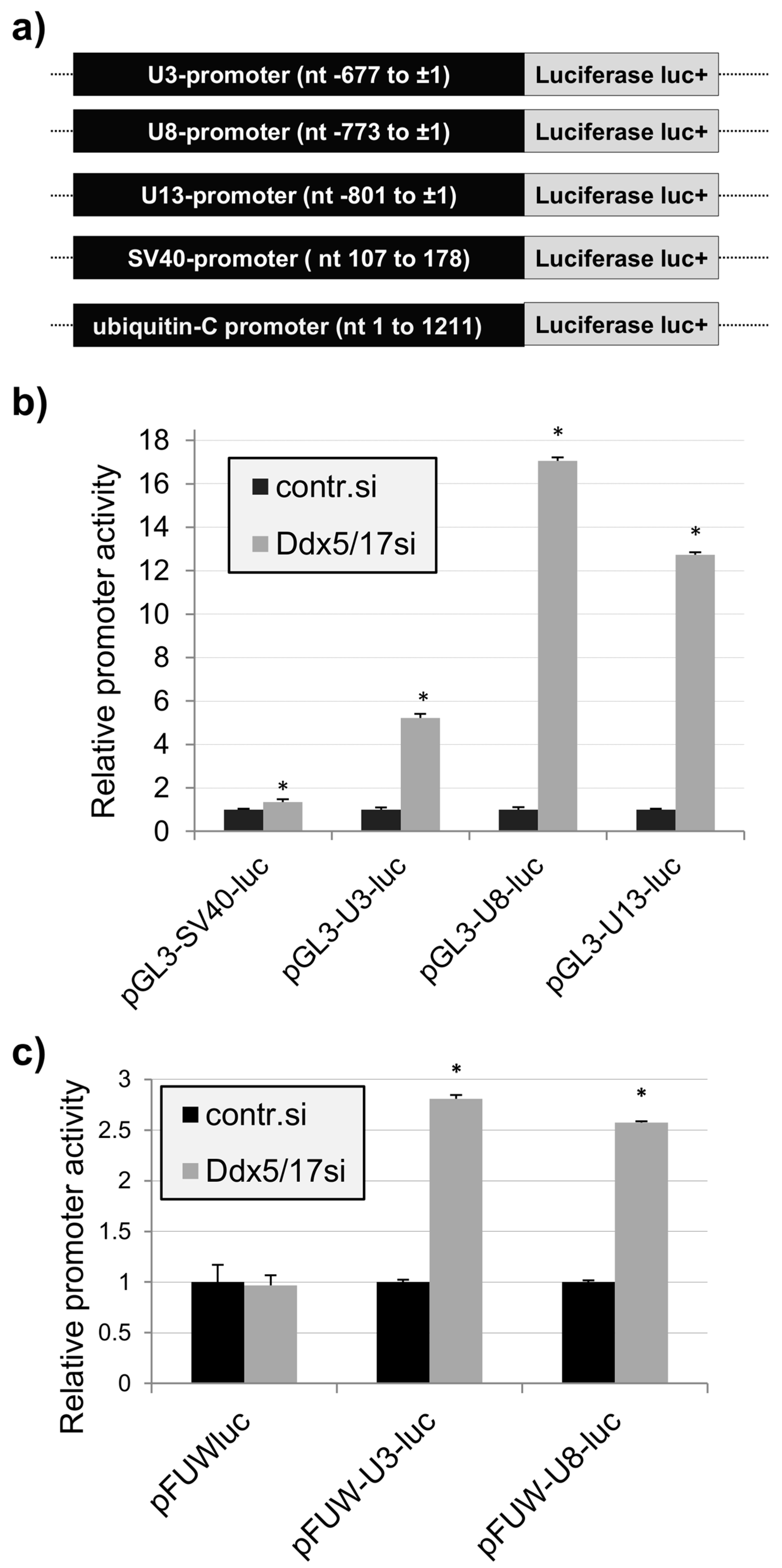
2.2. Promoter-Dependent Regulation of the U3-, U8-, and U13snoRNA Expression by Ddx5/Ddx17
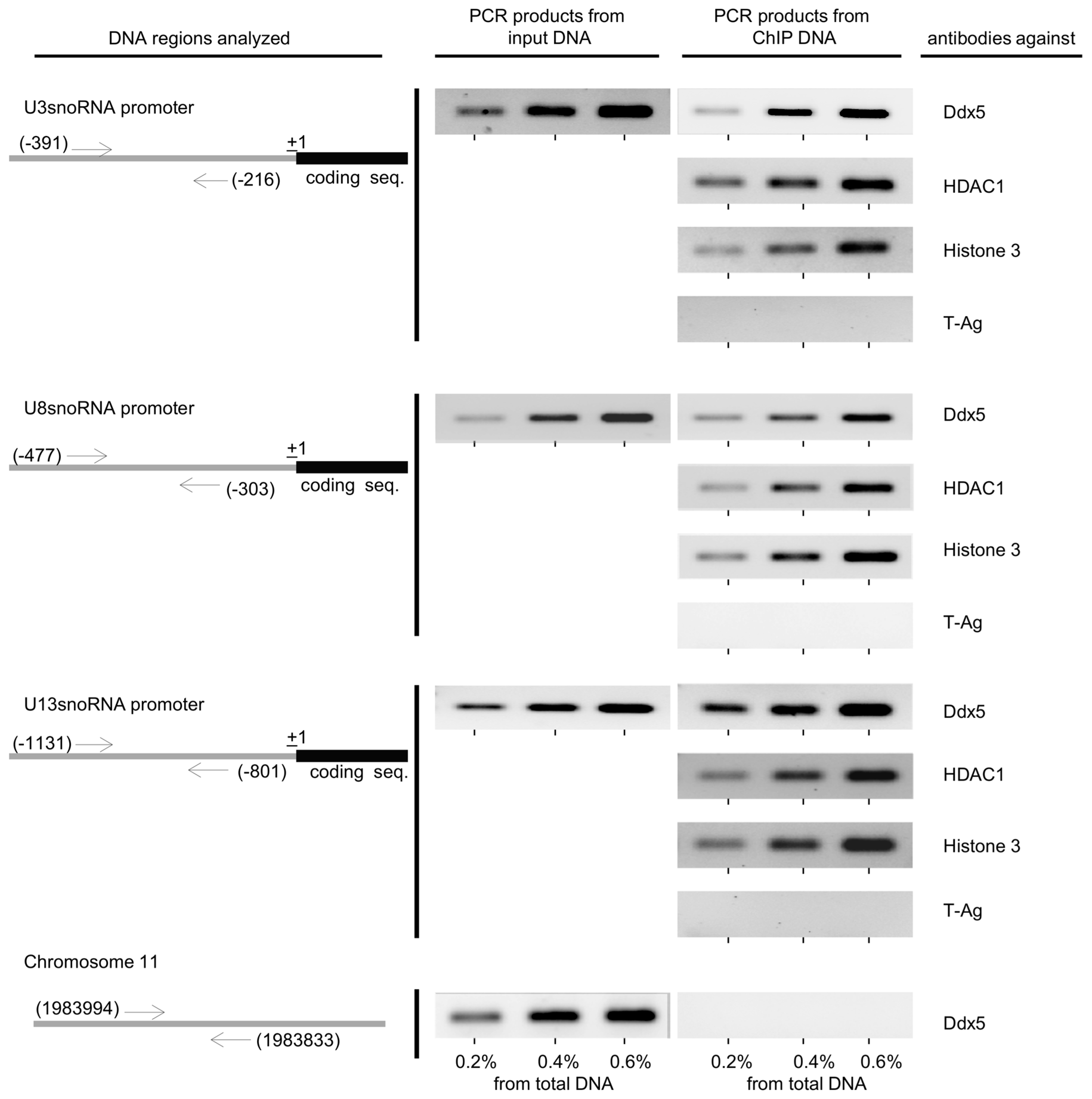
2.3. The Role of HDAC1 in the Regulation of U3-, U8-, and U13snoRNA Transcription
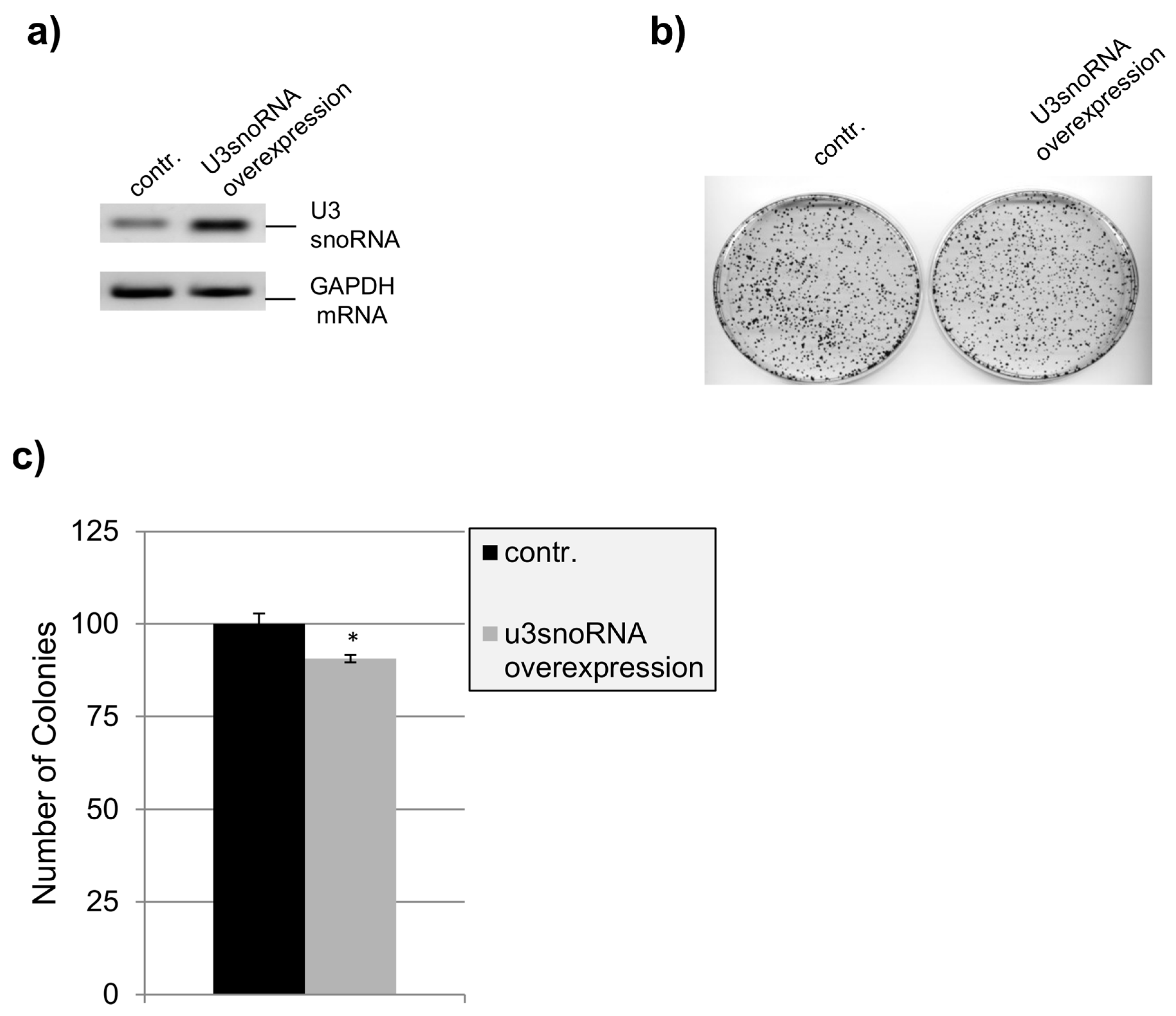
2.4. Influence of U3snoRNA Expression Level on Cell Proliferation
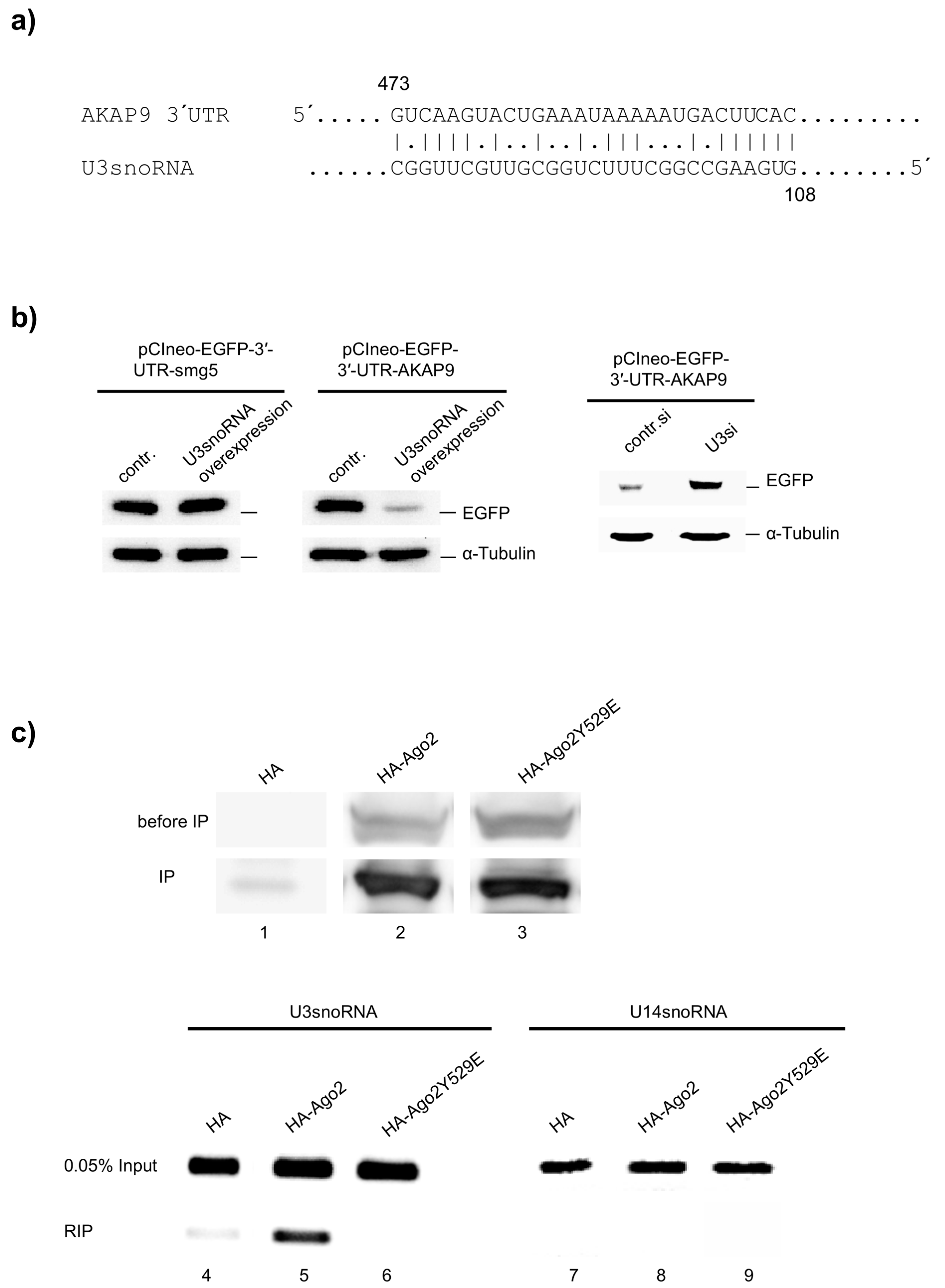
2.5. miRNA Function of U3snoRNA
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Cell Culture, Cell Transfection, and Exogenous Gene Expression
4.3. Lentiviral Gene Transfer
4.4. Reporter Assays
4.5. RNA Isolation
4.6. Northern Blot Analysis
4.7. Antibodies
4.8. RNA Immunoprecipitation (RIP)
4.9. Chromatin Immunoprecipitation (ChIP)
4.10. Cell Viability and Proliferation
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Maxwell, E.S.; Fournier, M.J. The small nucleolar RNAs. Annu. Rev. Biochem. 1995, 64, 897–934. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.P.; Omer, A. Small non-coding RNAs in Archaea. Curr. Opin. Microbiol. 2005, 8, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, L.B.; Steitz, J.A. Guided tours: From precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999, 11, 378–384. [Google Scholar] [CrossRef]
- Kiss, T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 2002, 109, 145–148. [Google Scholar] [CrossRef]
- Peculis, B.A.; Steitz, J.A. Disruption of U8 nucleolar snRNA inhibits 5.8s and 28s rRNA processing in the Xenopus oocyte. Cell 1993, 73, 1233–1245. [Google Scholar] [CrossRef]
- Sharma, K.; Tollervey, D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18s rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999, 19, 6012–6019. [Google Scholar] [CrossRef] [PubMed]
- Cavaille, J.; Hadjiolov, A.A.; Bachellerie, J.P. Processing of mammalian rRNA precursors at the 3′ end of 18s rRNA. Identification of cis-acting signals suggests the involvement of U13 small nucleolar RNA. Eur. J. Biochem. 1996, 242, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011, 14, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Kishore, S.; Khanna, A.; Zhang, Z.; Hui, J.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010, 19, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snoRNA with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Yamada, K.; Avolio, F.; Scott, M.S.; van Koningsbruggen, S.; Barton, G.J.; Lamond, A.I. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol. Biol. Cell 2010, 21, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Steitz, J.A.; Tycowski, K.T. Small RNA chaperones for ribosome biogenesis. Science 1995, 270, 1626–1627. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Shu, M.D.; Steitz, J.A. A mammalian gene with introns instead of exons generating stable RNA products. Nature 1996, 379, 464–466. [Google Scholar] [CrossRef] [PubMed]
- Tyc, K.; Steitz, J.A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989, 8, 3113–3119. [Google Scholar] [PubMed]
- Hirling, H.; Scheffner, M.; Restle, T.; Stahl, H. RNA helicase activity associated with the human p68 protein. Nature 1989, 339, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann-Schiffler, H.; Rossler, O.G.; Stahl, H. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J. Biol. Chem. 2002, 277, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Rossler, O.G.; Straka, A.; Stahl, H. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 2001, 29, 2088–2096. [Google Scholar] [CrossRef] [PubMed]
- Jalal, C.; Uhlmann-Schiffler, H.; Stahl, H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007, 35, 3590–3601. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J.; Bates, G.J.; Nicol, S.M.; Gregory, D.J.; Perkins, N.D.; Fuller-Pace, F.V. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol. Biol. 2004, 5, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wortham, N.C.; Ahamed, E.; Nicol, S.M.; Thomas, R.S.; Periyasamy, M.; Jiang, J.; Ochocka, A.M.; Shousha, S.; Huson, L.; Bray, S.E.; et al. The DEAD-box protein p72 regulates ERα-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERα-positive breast cancer. Oncogene 2009, 28, 4053–4064. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.P.; Liao, J.P.; Shen, J.; Yu, L.; Liu, B.L.; Liu, L.; Li, R.Y.; Ji, L.; Dorsey, S.G.; Jiang, Z.R.; et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012, 31, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Maragkakis, M.; Reczko, M.; Simossis, V.A.; Alexiou, P.; Papadopoulos, G.L.; Dalamagas, T.; Giannopoulos, G.; Goumas, G.; Koukis, E.; Kourtis, K.; et al. DIANA-microT web server: Elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009, 37, W273–W276. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.S.; Garchow, B.G.; Liu, X.; Yeung, J.; Morris, J.P.T.; Cuellar, T.L.; McManus, M.T.; Kiriakidou, M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009, 37, 7533–7545. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.X.; Postigo, A.A.; Dean, D.C. Rb interacts with histone deacetylase to repress transcription. Cell 1998, 92, 463–473. [Google Scholar] [CrossRef]
- Vanhaecke, T.; Papeleu, P.; Elaut, G.; Rogiers, V. Trichostatin A-like hydroxamate histone deacetylase inhibitors as therapeutic agents: Toxicological point of view. Curr. Med. Chem. 2004, 11, 1629–1643. [Google Scholar] [CrossRef] [PubMed]
- Hassig, C.A.; Tong, J.K.; Fleischer, T.C.; Owa, T.; Grable, P.G.; Ayer, D.E.; Schreiber, S.L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 1998, 95, 3519–3524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.W.; Iratni, R.; Erdjument-Bromage, H.; Tempst, P.; Hampsey, M.; Reinberg, D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1998, 1, 1021–1031. [Google Scholar] [CrossRef]
- Verheggen, C.; Lafontaine, D.L.; Samarsky, D.; Mouaikel, J.; Blanchard, J.M.; Bordonne, R.; Bertrand, E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002, 21, 2736–2745. [Google Scholar] [CrossRef] [PubMed]
- Kenmochi, N.; Higa, S.; Yoshihama, M.; Tanaka, T. U14 snoRNAs are encoded in introns of human ribosomal protein S13 gene. Biochem. Biophys. Res. Commun. 1996, 228, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Scott, M.S.; Yamada, K.; Avolio, F.; Barton, G.J.; Lamond, A.I. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011, 39, 3879–3891. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.S.; Avolio, F.; Ono, M.; Lamond, A.I.; Barton, G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009, 5, e1000507. [Google Scholar] [CrossRef] [PubMed]
- Flores-Jasso, C.F.; Arenas-Huertero, C.; Reyes, J.L.; Contreras-Cubas, C.; Covarrubias, A.; Vaca, L. First step in pre-miRNAs processing by human Dicer. Acta Pharmacol. Sin. 2009, 30, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Billy, E.; Brondani, V.; Zhang, H.; Muller, U.; Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 14428–14433. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Miyoshi, T.; Siomi, H. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol. Genet. Genom. 2010, 284, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef] [PubMed]
- Rudel, S.; Wang, Y.; Lenobel, R.; Korner, R.; Hsiao, H.H.; Urlaub, H.; Patel, D.; Meister, G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011, 39, 2330–2343. [Google Scholar] [CrossRef] [PubMed]
- Peters, L.; Meister, G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell 2007, 26, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Raska, I.; Ochs, R.L.; Andrade, L.E.; Chan, E.K.; Burlingame, R.; Peebles, C.; Gruol, D.; Tan, E.M. Association between the nucleolus and the coiled body. J. Struct. Biol. 1990, 104, 120–127. [Google Scholar] [CrossRef]
- Gerbi, S.A.; Borovjagin, A. U3 snoRNA may recycle through different compartments of the nucleolus. Chromosoma 1997, 105, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Darzacq, X.; Jady, B.E.; Verheggen, C.; Kiss, A.M.; Bertrand, E.; Kiss, T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002, 21, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Robb, G.B.; Brown, K.M.; Khurana, J.; Rana, T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005, 12, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Mattaloni, S.M.; Ferretti, A.C.; Tonucci, F.M.; Favre, C.; Goldenring, J.R.; Larocca, M.C. Centrosomal AKAP350 modulates the G1/S transition. Cell. Logist. 2013, 3, e26331. [Google Scholar] [CrossRef] [PubMed]
- Geissler, V.; Altmeyer, S.; Stein, B.; Uhlmann-Schiffler, H.; Stahl, H. The RNA helicase Ddx5/p68 binds to hUpf3 and enhances NMD of Ddx17/p72 and Smg5 mRNA. Nucleic Acids Res. 2013, 41, 7875–7888. [Google Scholar] [CrossRef] [PubMed]
- Boelz, S.; Neu-Yilik, G.; Gehring, N.H.; Hentze, M.W.; Kulozik, A.E. A chemiluminescence-based reporter system to monitor nonsense-mediated mRNA decay. Biochem. Biophys. Res. Commun. 2006, 349, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Muller, I.; Rossler, O.G.; Thiel, G. Pregnenolone sulfate activates basic region leucine zipper transcription factors in insulinoma cells: Role of voltage-gated Ca2+ channels and transient receptor potential melastatin 3 channels. Mol. Pharmacol. 2011, 80, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Stefano, L.; Al Sarraj, J.; Rossler, O.G.; Vinson, C.; Thiel, G. Up-regulation of tyrosine hydroxylase gene transcription by tetradecanoylphorbol acetate is mediated by the transcription factors Ets-like protein-1 (Elk-1) and Egr-1. J. Neurochem. 2006, 97, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Kaufmann, K.; Magin, A.; Lietz, M.; Bach, K.; Cramer, M. The human transcriptional repressor protein NAB1: Expression and biological activity. Biochim. Biophys. Acta 2000, 1493, 289–301. [Google Scholar] [CrossRef]
- Deppert, W.; Gurney, E.G.; Harrison, R.O. Monoclonal antibodies against simian virus 40 tumor antigens: Analysis of antigenic binding sites, using adenovirus type 2-simian virus 40 hybrid viruses. J. Virol. 1981, 37, 478–482. [Google Scholar] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismael, H.; Altmeyer, S.; Stahl, H. Regulation of the U3-, U8-, and U13snoRNA Expression by the DEAD Box Proteins Ddx5/Ddx17 with Consequences for Cell Proliferation and Survival. Non-Coding RNA 2016, 2, 11. https://doi.org/10.3390/ncrna2040011
Ismael H, Altmeyer S, Stahl H. Regulation of the U3-, U8-, and U13snoRNA Expression by the DEAD Box Proteins Ddx5/Ddx17 with Consequences for Cell Proliferation and Survival. Non-Coding RNA. 2016; 2(4):11. https://doi.org/10.3390/ncrna2040011
Chicago/Turabian StyleIsmael, Hala, Simone Altmeyer, and Hans Stahl. 2016. "Regulation of the U3-, U8-, and U13snoRNA Expression by the DEAD Box Proteins Ddx5/Ddx17 with Consequences for Cell Proliferation and Survival" Non-Coding RNA 2, no. 4: 11. https://doi.org/10.3390/ncrna2040011




